This repository contains the implementation of applications with PPML-Omics from:
Juexiao Zhou, Siyuan Chen, et al. "PPML-Omics: a Privacy-Preserving federated Machine Learning method protects patients’ privacy from omic data"
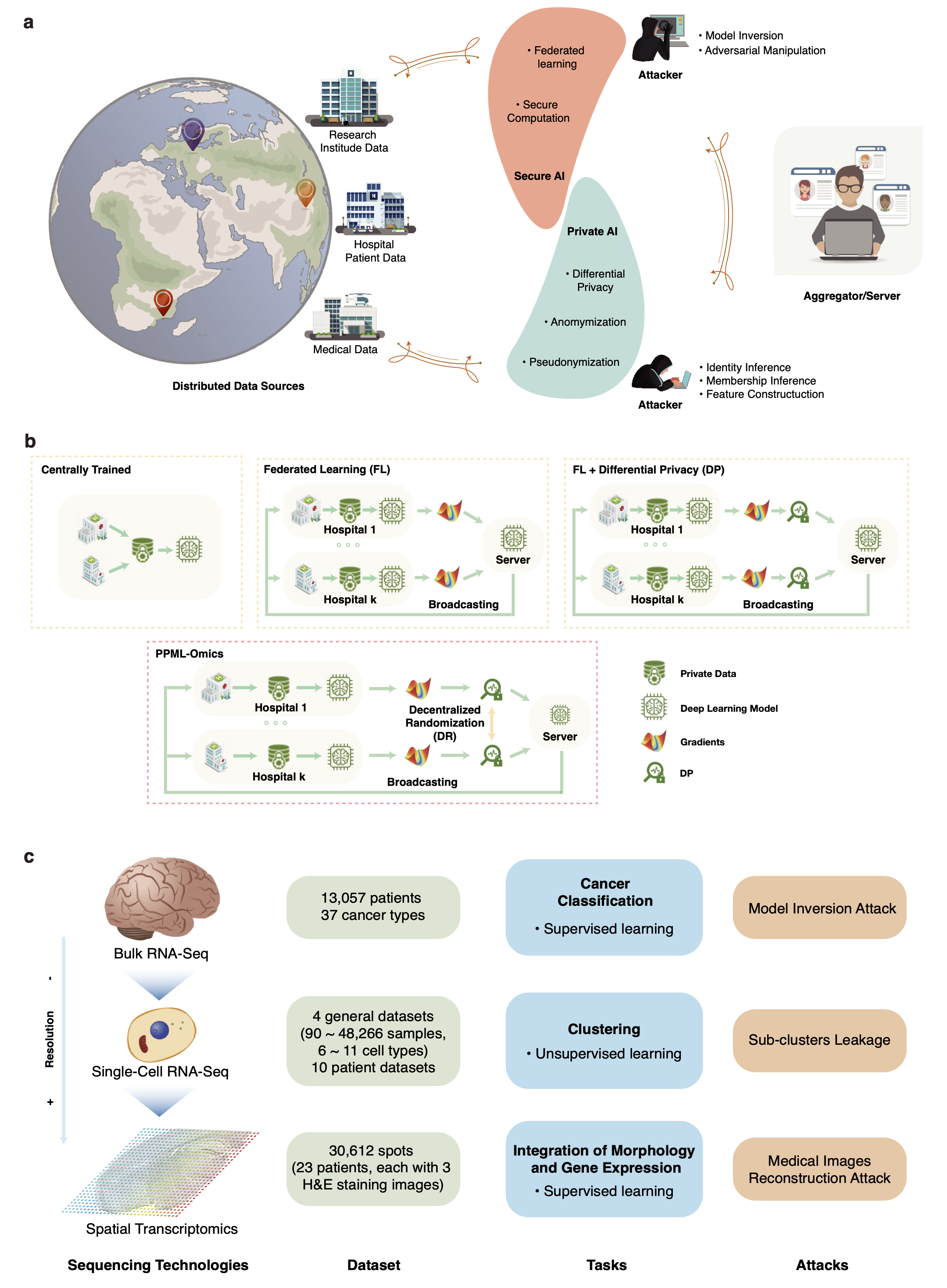
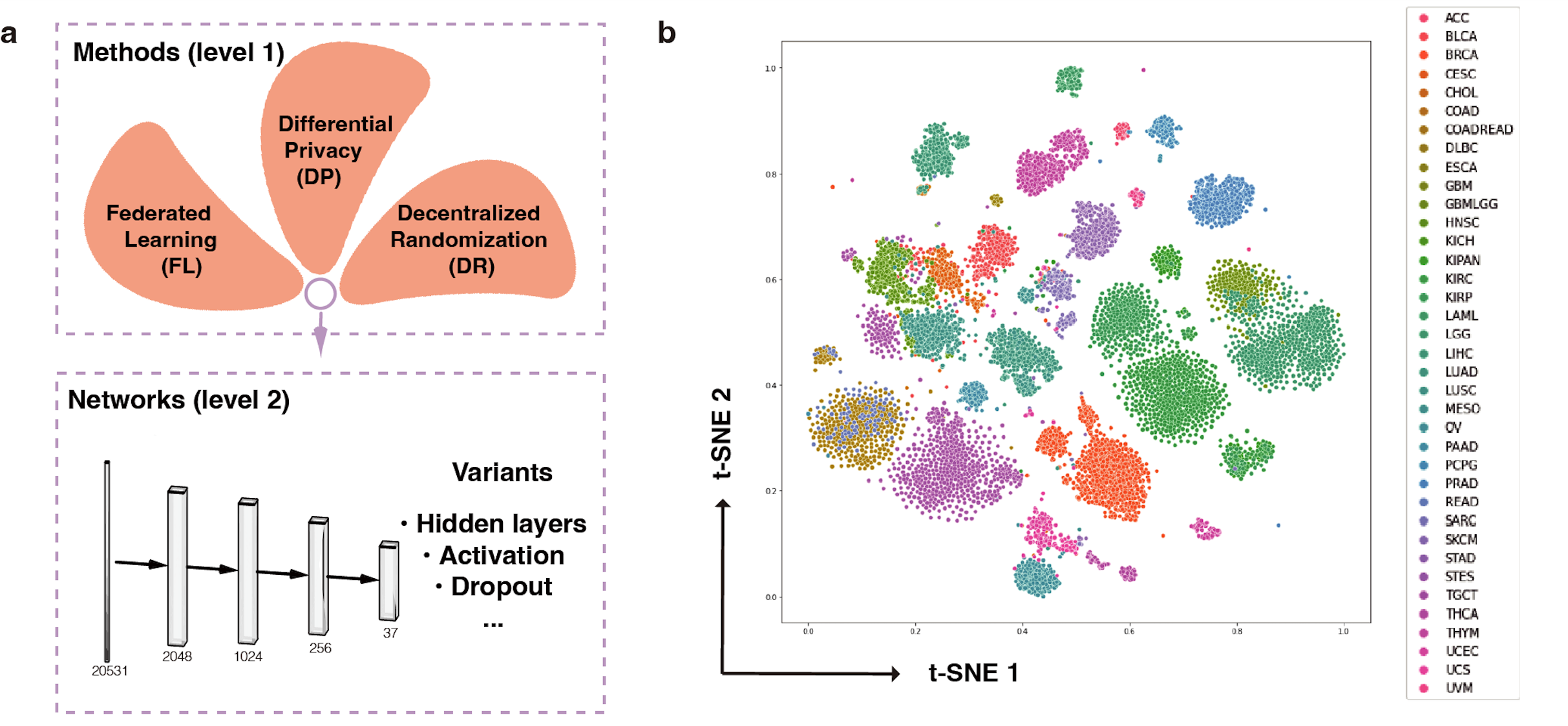
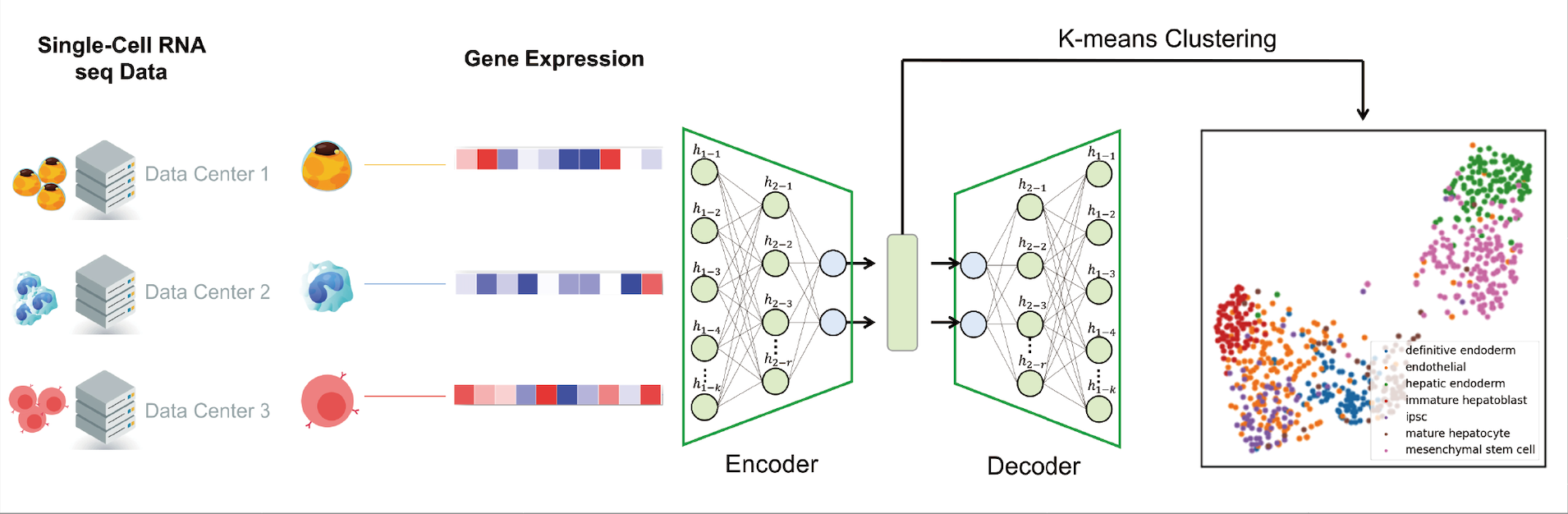
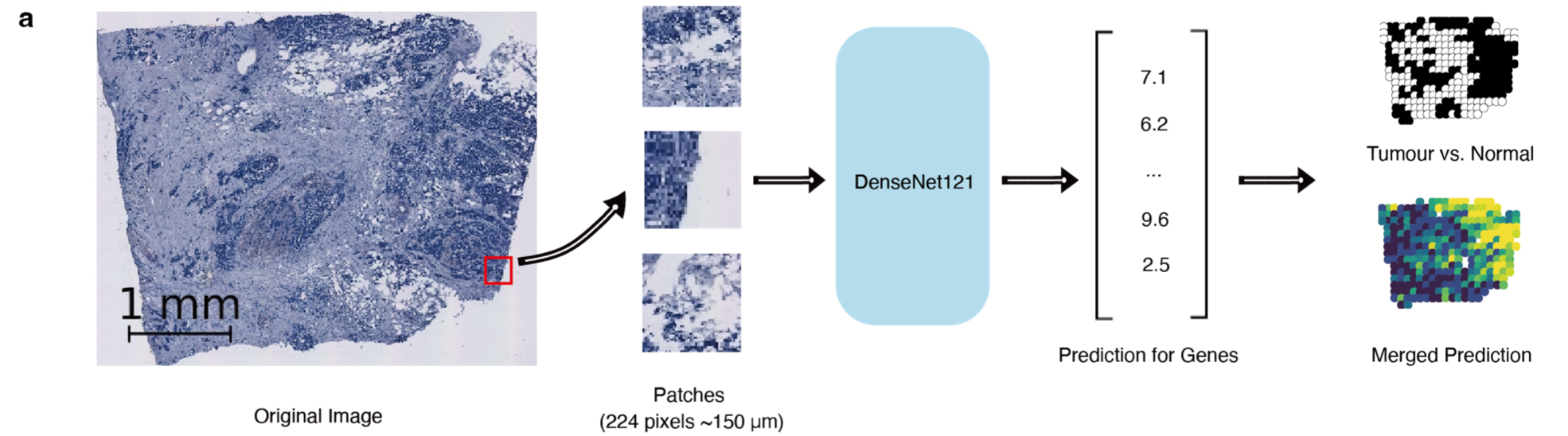
Fig. 1 PPML-Omics: a Privacy-Preserving federated Machine Learning method protects patients’ privacy from omic data. a, Schematic overview of the relationships and interactions between distributed data owners, aggregator, attackers and techniques in the field of secure and private AI. b, Schematic overview of different methods, including centrally trained method, federated learning (FL), FL with differential privacy (DP), and PPML-Omics. c, Illustration of 3 representative tasks, datasets and attacks of omic data in this paper for demonstrating the utility and privacy-preserving capability of PPML-Omics, including the 1) cancer classification with bulk RNA-seq, 2) clustering with scRNA-seq and 3) integration of morphology and gene expression with spatial transcriptomics.
/Attack: source codes for attack experiments
/Application: examples of applying PPML-Omics, users can eaisy modify based on it to meet their own requirements.
conda create -n ppmlomics python=3.9
conda activate ppmlomics
conda install mamba -y
mamba install pytorch torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia -y
mamba install seaborn -y
mamba install matplotlib -y
mamba install tqdm -y
mamba install scikit-learn -y
mamba install -c conda-forge scipy -y
mamba install numpy=1.20.3 -y
mamba install -c conda-forge umap-learn -y
mamba install -c conda-forge openslide-python -y
pip install tenseal
Please run all codes in this section with Application/CancerTypeClassification as root.
The complete dataset can be downloaded via Google Drive.
python 01.processData.py
python 02.simulationApp.py --help
usage: 02.simulationApp.py [-h] [--device DEVICE] [--epochs EPOCHS] [--batch_size BATCH_SIZE] [--lr LR] [--epsilon EPSILON] [--delta DELTA] [--mode MODE] [--client CLIENT]
[--l2_clip L2_CLIP] [--nprocess NPROCESS] [--expname EXPNAME] [--train_data TRAIN_DATA] [--test_data TEST_DATA] [--shuffle_model SHUFFLE_MODEL]
dfsa
optional arguments:
-h, --help show this help message and exit
--device DEVICE default: cuda:0
--epochs EPOCHS default: 50
--batch_size BATCH_SIZE
default: 1
--lr LR default: 0.01
--epsilon EPSILON default: 100
--delta DELTA default: 0.5
--mode MODE default: SGD, DP, PPMLOmics
--client CLIENT default: 3
--l2_clip L2_CLIP default: 5
--nprocess NPROCESS default: 20
--expname EXPNAME experiment name
--train_data TRAIN_DATA
will load data/{}.npy
--test_data TEST_DATA
will load data/{}.npy
--shuffle_model SHUFFLE_MODEL
0: off, 1: on
PPML-Omics provides 4 basic modes for calculate the gradients: SGD, SIGNSGD, DP, DPSIGNSGD, and 1 shuffling mode, with --shuffle_model sets to 1, which can be freely combined into different systems.
python 02.simulationApp.py --mode SGD --client 1 --epochs 10 --batch_size 8 --lr 0.001 --expname PureSGD0 --train_data train_log10_0 --test_data test_log10_0
python 02.simulationApp.py --mode SGD --client 5 --epochs 10 --batch_size 8 --lr 0.001 --expname FL --train_data train_log10_0 --test_data test_log10_0
for e in 50 40 30 20 10 5 1
do
python 02.simulationApp.py --mode DP --client 20 --epochs 10 --device cuda:1 --batch_size 8 --lr 0.001 --epsilon ${e} --expname DP_e${e} --train_data train_log10_0 --test_data test_log10_0
done
for e in 50 40 30 20 10 5 1
do
python 02.simulationApp.py --mode PPMLOmics --client 20 --epochs 10 --batch_size 8 --lr 0.001 --epsilon ${e} --expname PPMLOmics_e${e} --train_data train_log10_0 --test_data test_log10_0
done
for cli in 5 10 20 50
do
python 02.simulationApp.py --mode PPMLOmics --client ${cli} --epochs 10 --device cuda:1 --batch_size 8 --lr 0.001 --epsilon 20 --expname PPMLOmics_e20_cli${cli} --train_data train_log10_0 --test_data test_log10_0
done
python 03.attackApp.py --model path/to/model
MIAVisualization.ipynb
Please run all codes in this section with Application/SingleCellClustering as root.
The count data for 4 scRNA-seq dataset can be downloaded via Google Drive.
The original .rds file can be found in scDHA website
python 01.simulationApp.py --help
usage: 01.simulationApp.py [-h] [--device DEVICE] [--epochs EPOCHS] [--batch_size BATCH_SIZE] [--lr LR] [--epsilon EPSILON] [--delta DELTA] [--mode MODE] [--client CLIENT] [--l2_clip L2_CLIP] [--nprocess NPROCESS] [--dataset DATASET] [--expname EXPNAME] [--train_data TRAIN_DATA] [--test_data TEST_DATA] [--shuffle_model SHUFFLE_MODEL]
dfsa
optional arguments:
-h, --help show this help message and exit
--device DEVICE default: cuda:0
--epochs EPOCHS default: 50
--batch_size BATCH_SIZE
default: 1
--lr LR default: 0.01
--epsilon EPSILON default: 100
--delta DELTA default: 0.5
--mode MODE default: SGD, DP, PPMLOmics
--client CLIENT default: 3
--l2_clip L2_CLIP default: 5
--nprocess NPROCESS default: 20
--dataset DATASET dataset name
--expname EXPNAME experiment name
--train_data TRAIN_DATA
will load data/{}.npy
--test_data TEST_DATA
will load data/{}.npy
--shuffle_model SHUFFLE_MODEL
0: off, 1: on
for dataset in hrvatin pollen yan
do
python 01.simulationApp.py --dataset=$dataset --mode=SGD --client=1 --epochs=5 --batch_size=8 --lr=0.001 --expname="PureSGD"
done
for dataset in hrvatin pollen yan
do
python 01.simulationApp.py --dataset=$dataset --mode=SGD --client=5 --epochs=5 --batch_size=8 --lr=0.001 --expname="FL"
done
for dataset in hrvatin pollen yan
do
for e in 2 3 5 10 20 50
do
python 01.simulationApp.py --dataset=$dataset --mode=DP --client=30 --epochs=5 --rep=3 --batch_size=8 --lr=0.001 --epsilon=${e} --delta=1e-5 --expname="DP"
done
done
for dataset in hrvatin pollen yan
do
for e in 2 3 5 10 20 50
do
python 01.simulationApp.py --dataset=$dataset --mode=PPMLOmics --client=30 --epochs=5 --rep=3 --batch_size=8 --lr=0.001 --epsilon=${e} --delta=1e-5 --expname="PPML-Omics"
done
done
for dataset in P0410 P1026
do
for e in 2 3 5 10 20 50
do
python 01.simulationApp.py --dataset=$dataset --mode=PPMLOmics --client=1 --epochs=5 --rep=3 --batch_size=8 --lr=0.001 --epsilon=${e} --delta=1e-5 --expname="PPML-Omics"
done
done
# For datasets: yan pollen hrvatin
python 02.Test.py --dataset="yan" --model="model/PPMLOmics_yan_modelbest.tar" --expname="PPMLOmics_e5"
# For patients
python 04.TestPatient.py --dataset="P0123" --model="model/PPMLOmics_P0123_epsilon_5.0_modelbest.tar" --expname="PPMLOmics_e5"
Please run all codes in this section with Application/SpatialTranscriptomics as root.
The complete dataset can be downloaded via Google Drive.
python 01.simulationApp.py --help
usage: 01.simulationApp.py [-h] [--device DEVICE] [--epochs EPOCHS] [--batch_size BATCH_SIZE] [--lr LR] [--epsilon EPSILON] [--delta DELTA] [--mode MODE] [--client CLIENT] [--l2_clip L2_CLIP] [--nprocess NPROCESS] [--expname EXPNAME] [--shuffle_model SHUFFLE_MODEL]
dfsa
optional arguments:
-h, --help show this help message and exit
--device DEVICE default: cuda:0
--epochs EPOCHS default: 50
--batch_size BATCH_SIZE
default: 1
--lr LR default: 0.01
--epsilon EPSILON default: 100
--delta DELTA default: 0.5
--mode MODE default: SGD, DP, PPMLOmics
--client CLIENT default: 3
--l2_clip L2_CLIP default: 5
--nprocess NPROCESS default: 20
--expname EXPNAME experiment name
--shuffle_model SHUFFLE_MODEL
0: off, 1: on
python 01.simulationApp.py --device cuda:0 --mode SGD --client 1 --epochs 10 --batch_size 32 --nprocess 40 --lr 1e-6 --expname PureSGD
python 01.simulationApp.py --device cuda:1 --mode SGD --client 30 --epochs 10 --batch_size 32 --nprocess 40 --lr 1e-6 --expname FL
for e in 10 5 1 0.5 0.1
do
python 01.simulationApp.py --device cuda:1 --mode DP --client 30 --epochs 10 --epsilon ${e} --lr 1e-5 --delta 1e-5 --batch_size 32 --nprocess 40 --l2_clip 20 --expname DP_e${e}
done
for e in 10 5 1 0.5 0.1
do
python 01.simulationApp.py --device cuda:0 --mode PPMLOmics --client 30 --epochs 10 --epsilon ${e} --lr 1e-6 --delta 1e-5 --batch_size 32 --nprocess 40 --l2_clip 20 --expname PPMLOmics_e${e}
done
python 02.attackApp.py --mode SGD --expname iDLG_attack_SGD
python 02.attackApp.py --mode DP --expname iDLG_attack_DP --epsilon 0.01
If you find our work helpful, please cite our paper:
@article{zhou2024ppml,
title={PPML-Omics: A privacy-preserving federated machine learning method protects patients’ privacy in omic data},
author={Zhou, Juexiao and Chen, Siyuan and Wu, Yulian and Li, Haoyang and Zhang, Bin and Zhou, Longxi and Hu, Yan and Xiang, Zihang and Li, Zhongxiao and Chen, Ningning and others},
journal={Science Advances},
volume={10},
number={5},
pages={eadh8601},
year={2024},
publisher={American Association for the Advancement of Science}
}