Comments (6)
from ripser.py.
from ripser.py.
Hi Chris,
Thank you very much for your quick response - I will try it (we have previously used 1e-6 instead of zero, and it only worked so-so). Do you mind sharing what approaches you use to represent trajectories when passing them to ripser.py?
Thank you very much - best,
Wolfgang
from ripser.py.
from ripser.py.
Hi both,
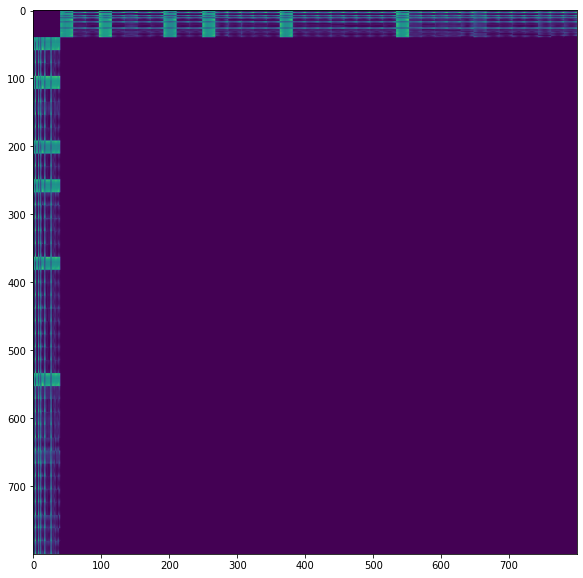
I have encountered a similar issue with the missing entries in a sparse matrix being interpreted as inf instead of a zero.
I generated a distance matrix that has a large zero block:
The dense matrix produces the correct results at the cost of storage and probably additional computation.
I'd like to add myself as another future user of the feature to optionally to treat the undefined elements of a sparse matrix as zero.
Best,
Vladimir
from ripser.py.
@wxmerkt a bit late to the party here but, in my experience, things work out as I think you would like them to in ripser.py if you explicitly store zeros in your sparse matrices (this can be done in a number of scipy sparse formats, though not in all). I can show you an example if you like (perhaps this got solved in the meantime?).
When making pyflagser (docs), we had to face some similar conundrums concerning the expected format of sparse matrices. In the end, we settled for a design choice which is explained in the function flagser_weighted -- analogous to ripser (it computes the same persistence diagrams when directed=False is passed!). In brief, you can pass sparse adjacency matrices with explicitly stored zeros and they are treated as zero filtration parameters, not as absent edges. The absent edges, as @ctralie pointed out is the case also in ripser.py, are the non-stored entries in the sparse matrix (the "sparse zeros", if you will). But again, I think ripser.py does the same thing!
from ripser.py.
Related Issues (20)
- Build failure on GNU/Linux (GCC 11.1.0 and Python 3.9.6 on Arch Linux) HOT 3
- Jupyter notebook crash on input of large sparse distance matrix HOT 1
- [Suggestion] Throw exception when computing bottleneck distance
- Dead comments in 0-dimensional computation/preferential treatment of edges with zero value
- get_greedy_perm (Samples)
- Get the indices of the vectors forming a hole HOT 2
- Install Ripser on ArchLinux HOT 2
- ld: cannot find /lib64/libpthread.so.0
- `ValueError: numpy.ndarray size changed` HOT 5
- Failed to build ripser, error: subprocess-exited-with-error HOT 6
- Knowing required RAM to run ripser
- Unsuccessful installation HOT 2
- Is there a way to get ripser to use virtual memory? HOT 2
- Failed building wheel for Ripser on Win11 HOT 3
- ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 96 from C header, got 88 from PyObject HOT 1
- Failed building wheel for ripser HOT 1
- Failing to generate H1 result with small dataset. HOT 1
- A potential issue with the `lower_star_img` function with negative numbers HOT 9
- Missing wheels for 0.6.5 HOT 5
- Fix failing docs
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.

from ripser.py.