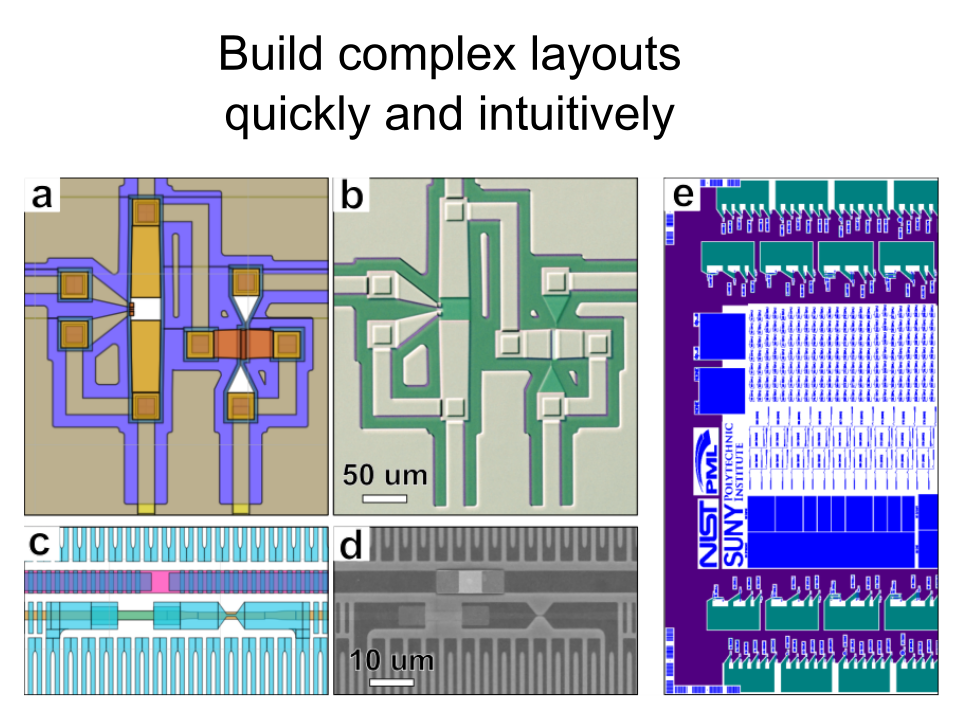
GDS scripting for Python that's intuitive, fast, and powerful.
- Installation / requirements
- Tutorial + examples (or try an interactive notebook)
- Geometry library + function documentation
- Changelog (latest update 1.7.2 on July 3, 2024)
- New KLayout-based boolean/offset/outline functions! These are under the name
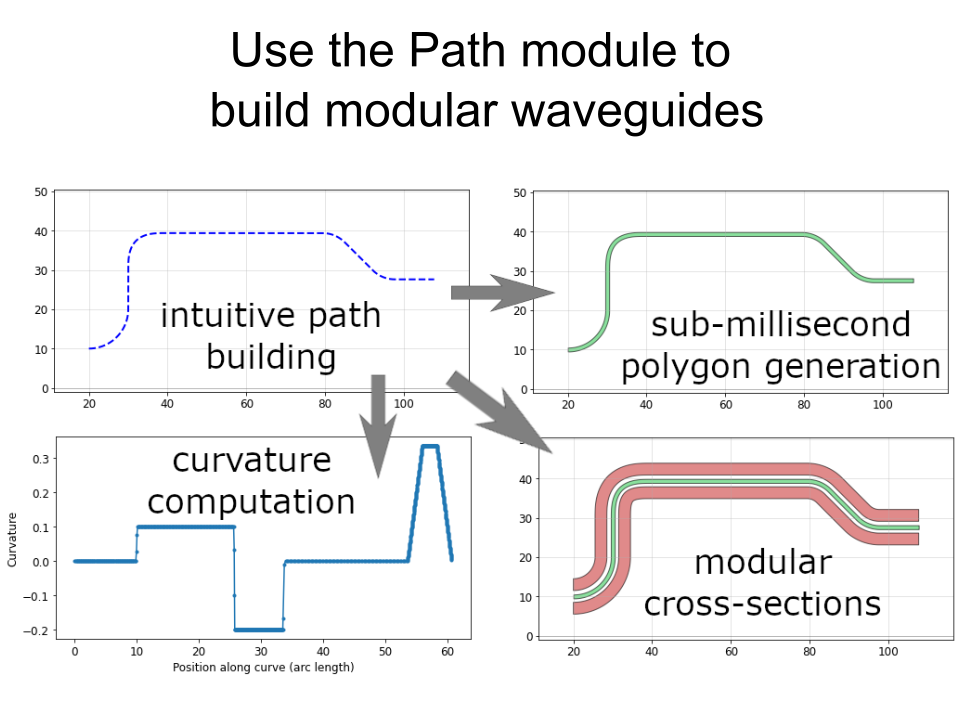
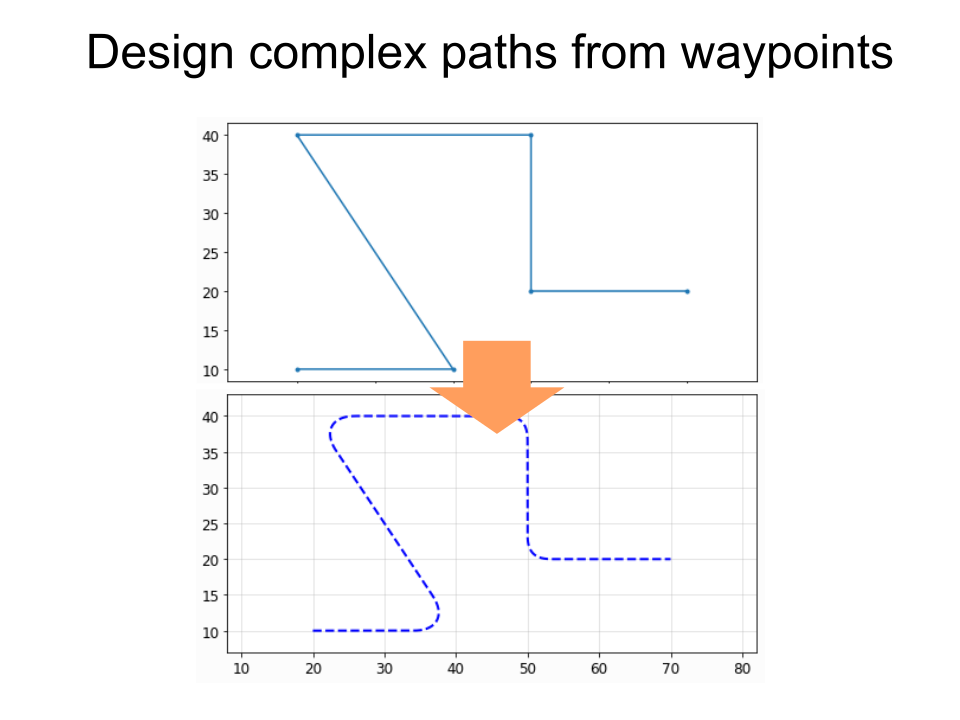
pg.kl_boolean(),pg.kl_offset,pg.kl_outline(),pg.kl_invert(). They utilize the excellent KLayout tile processor, which allows breaking down & parallelizing these operations--in a nutshell, these operations should be much, much faster, and they also are more robust than the gdspy/clipper implementation. To use these new functions, you must firstpip install klayout - Path.interpolate() now allows easy placement of objects alongside a path (e.g. for placing vias). See the tutorial for more information
- New KLayout-based boolean/offset/outline functions! These are under the name
If you found PHIDL useful, please consider citing it in (just one!) of your publications -- we appreciate it greatly. (BibTeX)
- McCaughan, A. N., et. al. PHIDL: Python-based layout and geometry creation for nanolithography. J. Vac. Sci. Technol. B 39, 062601 (2021). http://dx.doi.org/10.1116/6.0001203
- Install or upgrade with
pip install -U phidl - Install with
pip install -U phidl[all]to include optional dependencies (e.g. freetype-py, klayout, rectpack) - Python version >=3.6
- Install with test dependencies with
pip install -U phidl[test](includesallextras as well) - Run tests with
pytest(orpython -m pytest)
fiddle (verb) - /ˈfidl/ - to make minor manual movements, especially to adjust something
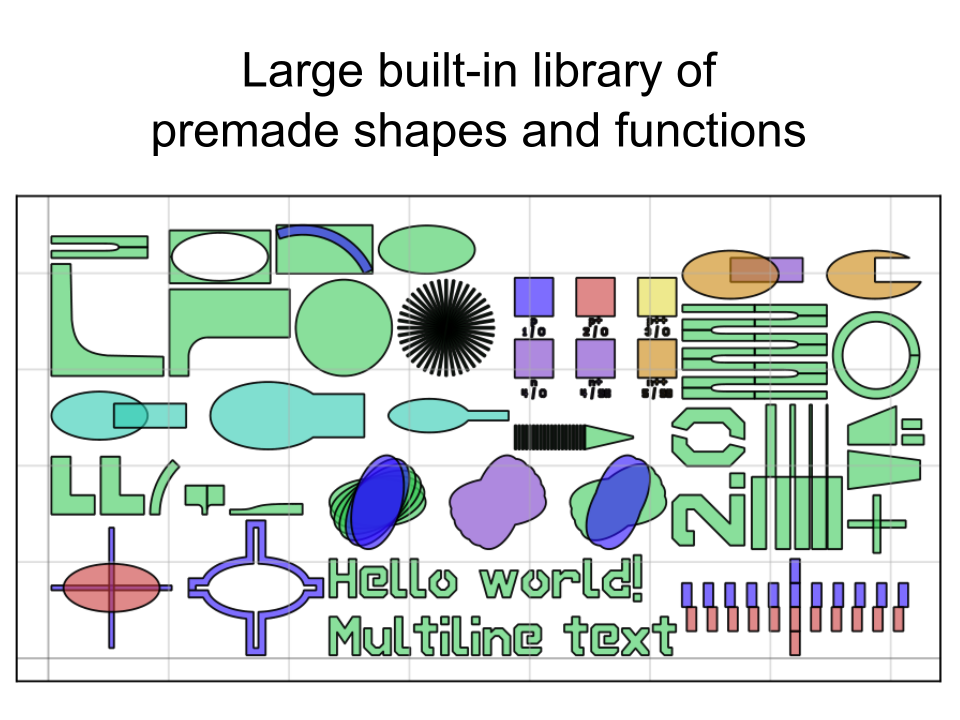
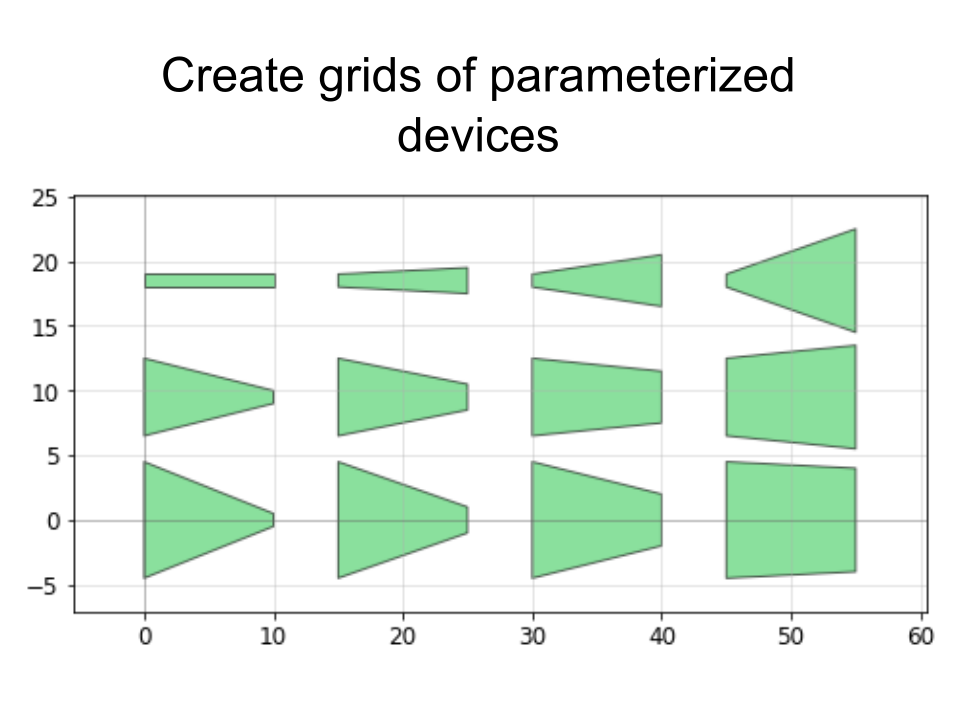
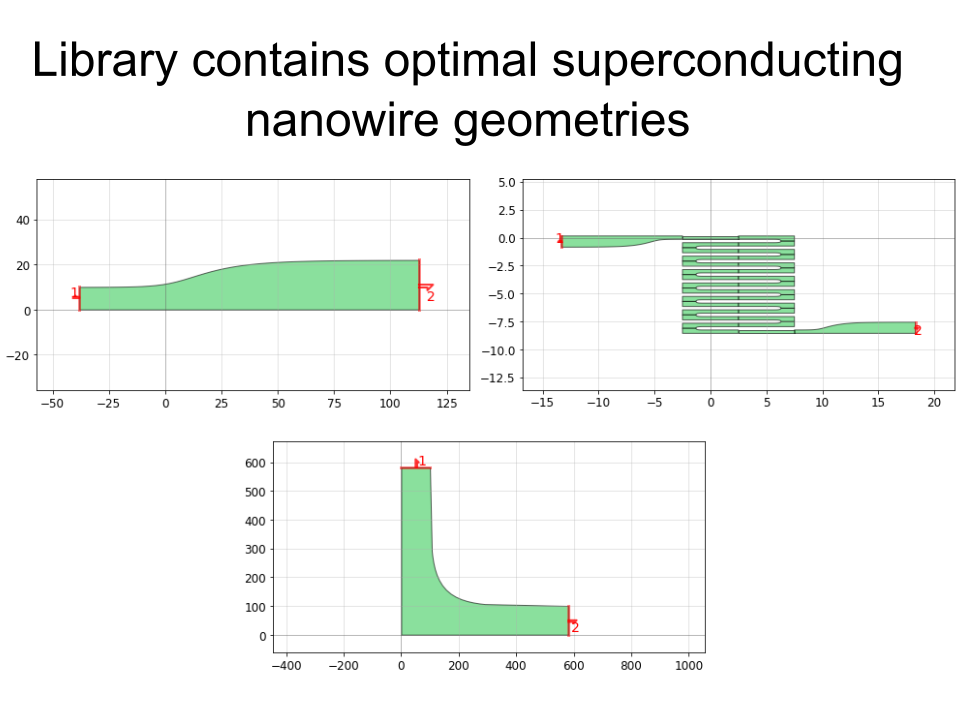
PHIDL is an open-source GDS-based CAD tool for Python that significantly extends the excellent gdspy. The base installation includes a large library of simple shapes (e.g. rectangles, circles), photonic structures (e.g. sine curve waveguides), and superconducting nanowire shapes (e.g. single photon detectors) that are fully parameterized. It also has a built-in quick-plotting function based on matplotlib (or Qt) that allows you view the state of any GDS object, useful when scripting geometry-making functions. It also has a geometry library reference and a set of very thorough tutorials that will walk you through the process of getting acquainted with PHIDL.
The goal is to bring the usability of Illustrator / Inkscape drawing programs to the GDS scripting world. Like Python itself, it aims to be readable, and intuitive. For instance, when building a geometry you don't have to worry about what the exact coordinates are anymore. If you want to separate two ellipses in the x direction by 5 units, you can do things like this:
ellipse1.xmin = ellipse2.xmax + 5
or if you want to move then rotate one ellipse by 45 degrees you can do
ellipse2.move([1,7]).rotate(45)
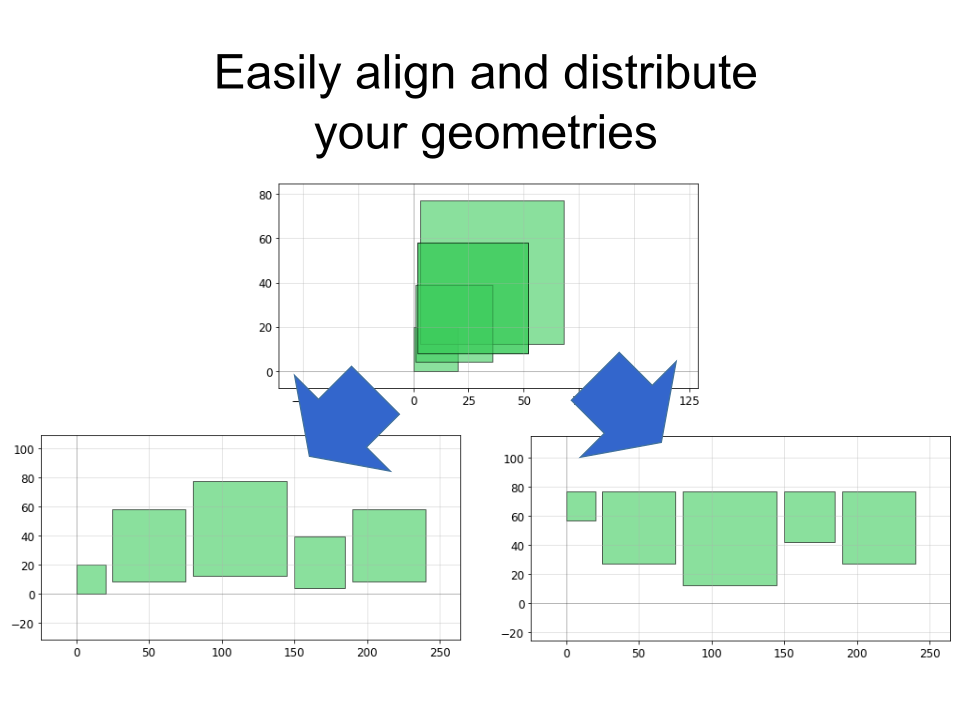
There's a few dozen shortcuts like this that make life easier built into PHIDL--they're simple, but they make a world of difference when you just want to e.g. space a ring resonator some distance from a waveguide without having to track each and every coordinate of the shape.
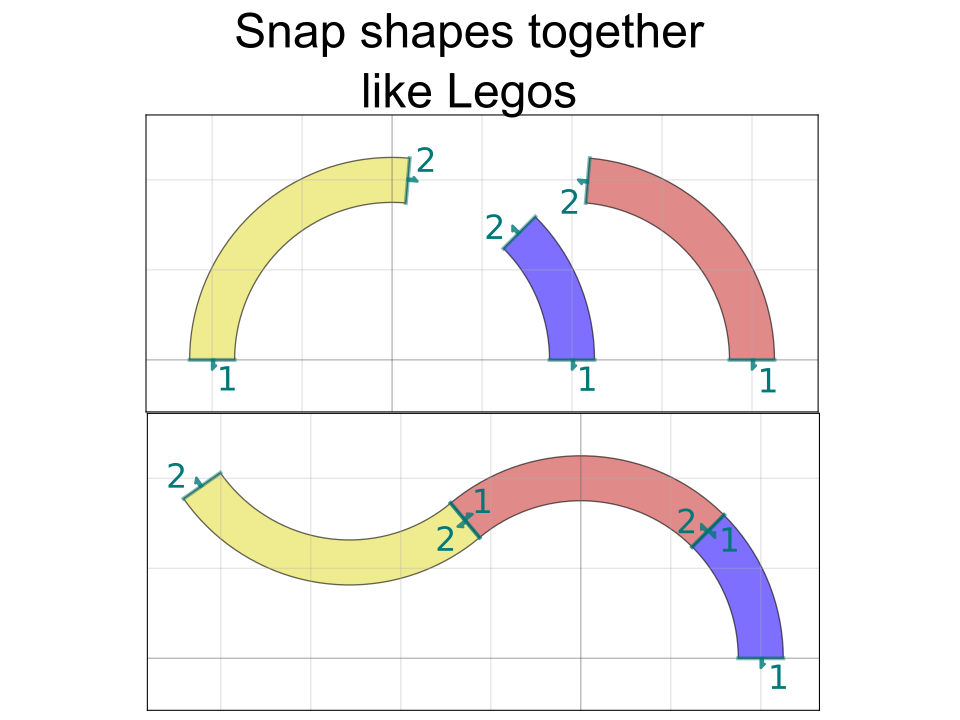
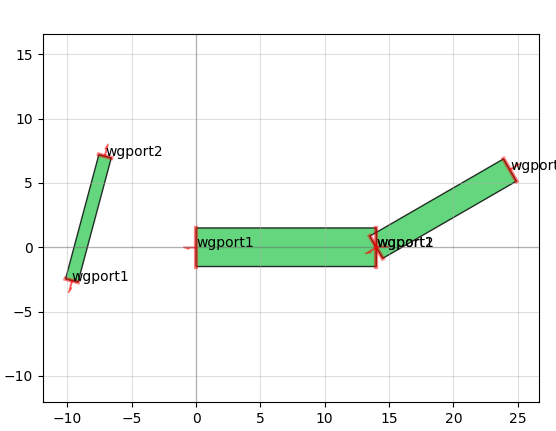
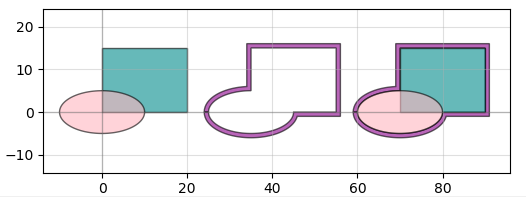
There's also a "port" functionality that allows you to snap together geometry like Legos without caring about where exactly the absolute coordinates of either geometry is. For instance, connecting the above misaligned rectangles is a two-line command:
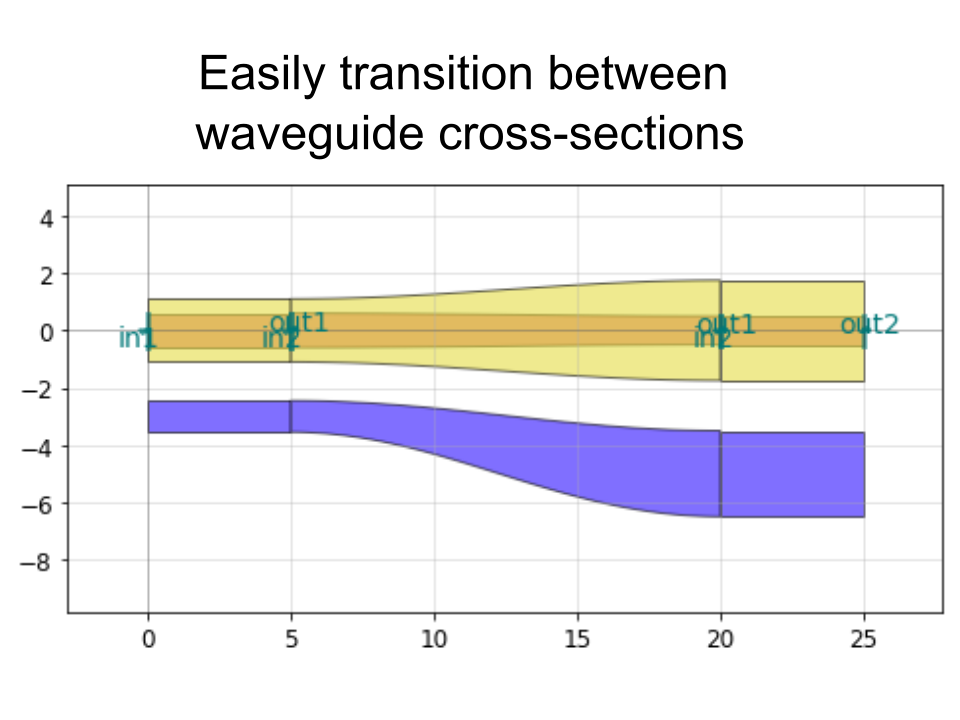
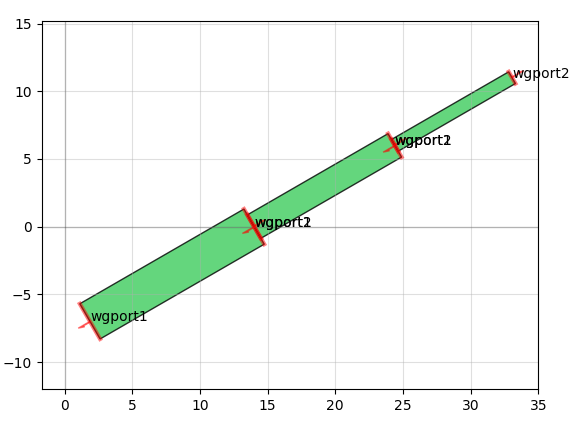
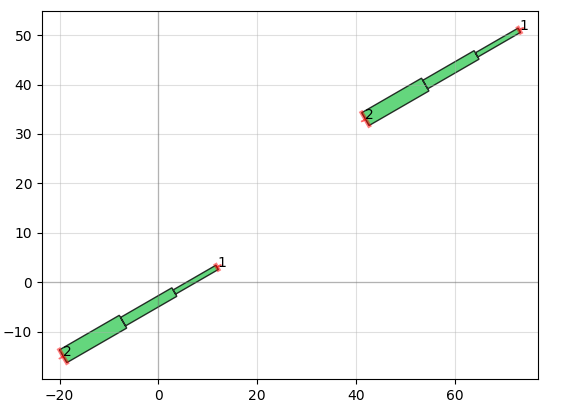
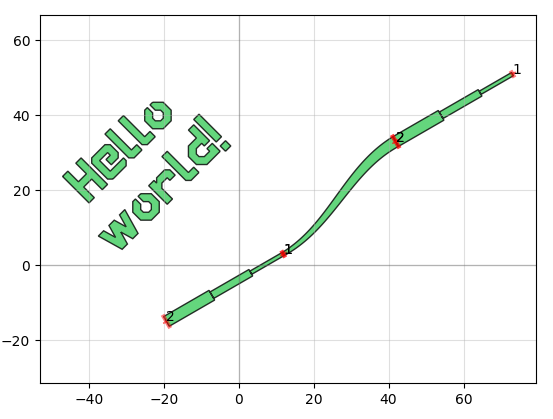
It also allows you to do things like add text and create smooth or straight routing curves between "ports" of different devices, convenient for making electrical or optical connections:
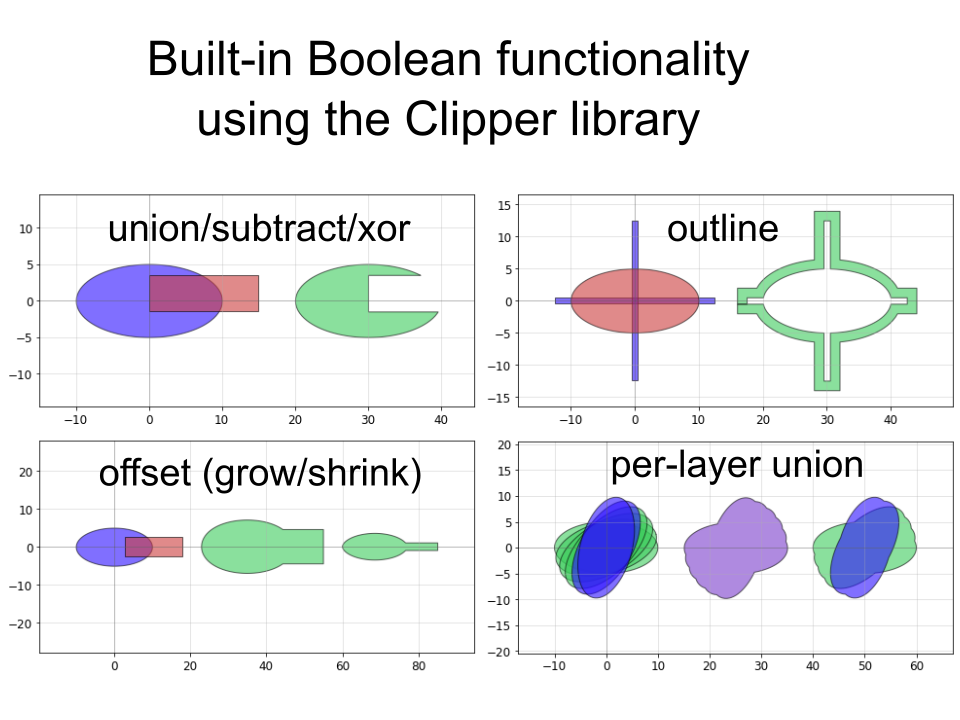
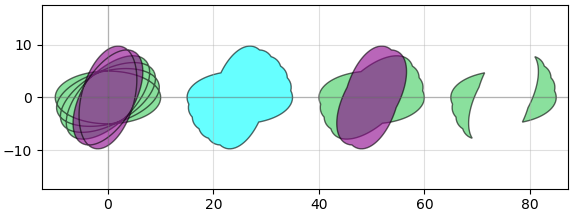
Other useful functionality available are standard operations like booleans:
and less standard ones like creating outlines. A whole layout can be outlined directly in the GDS without requiring you to use Beamer (useful for positive-tone resist structures):
pg.outline(D, distance = 0.7, layer = 4)
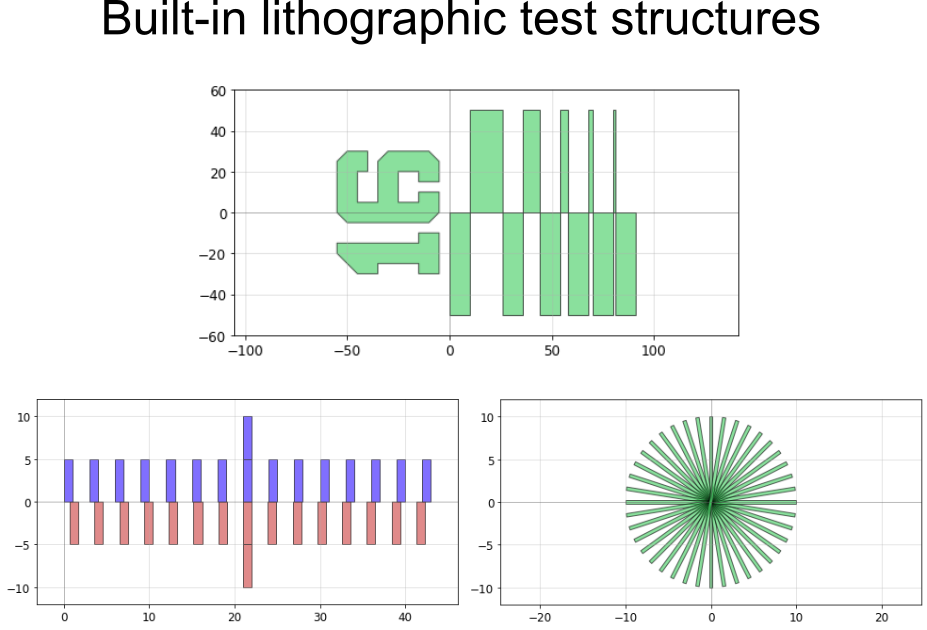
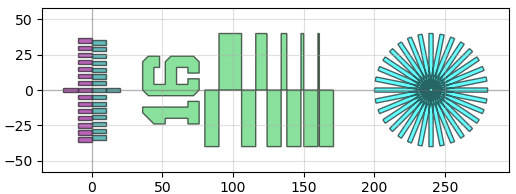
The geometry library also has useful resolution test-structures built into it, for instance
pg.litho_calipers(num_notches = 7, offset_per_notch = 0.1)
pg.litho_steps(line_widths = [1,2,4,8,16])
pg.litho_star(num_lines = 16, line_width = 3)
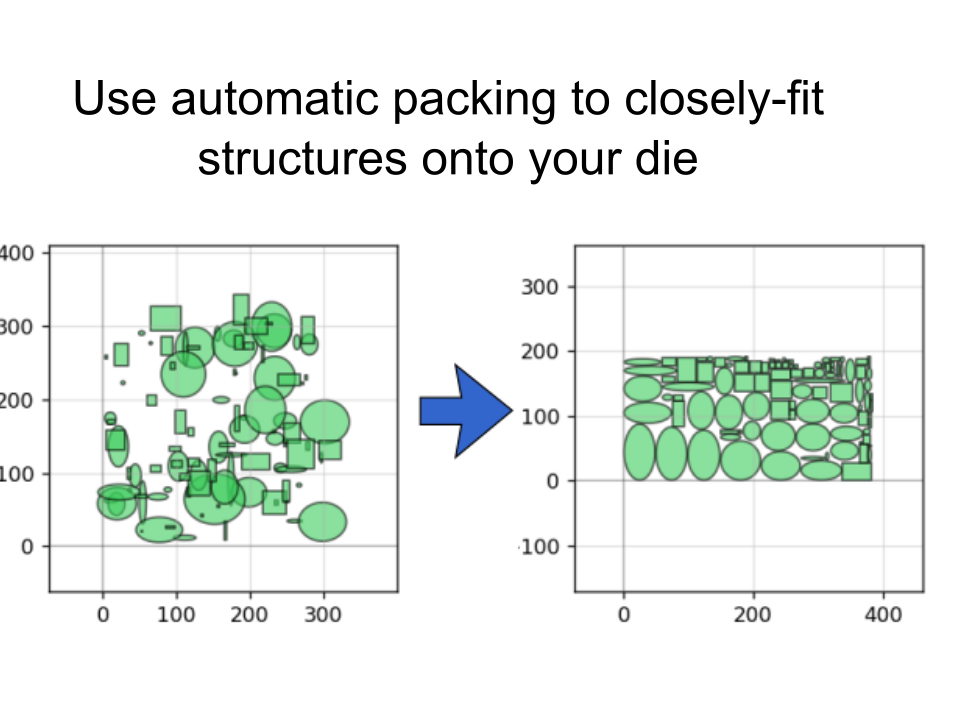
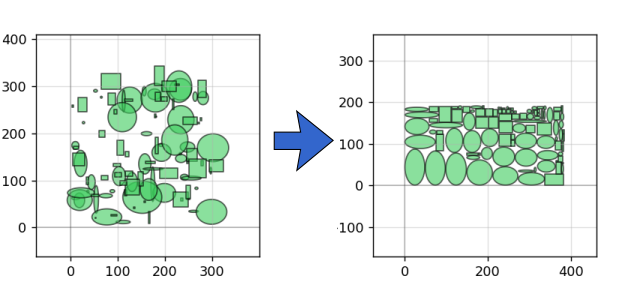
There are also handy functions to help pack shapes into as small an area as possible:
pg.packer(D_list, spacing = 1.25, aspect_ratio = (2,1))