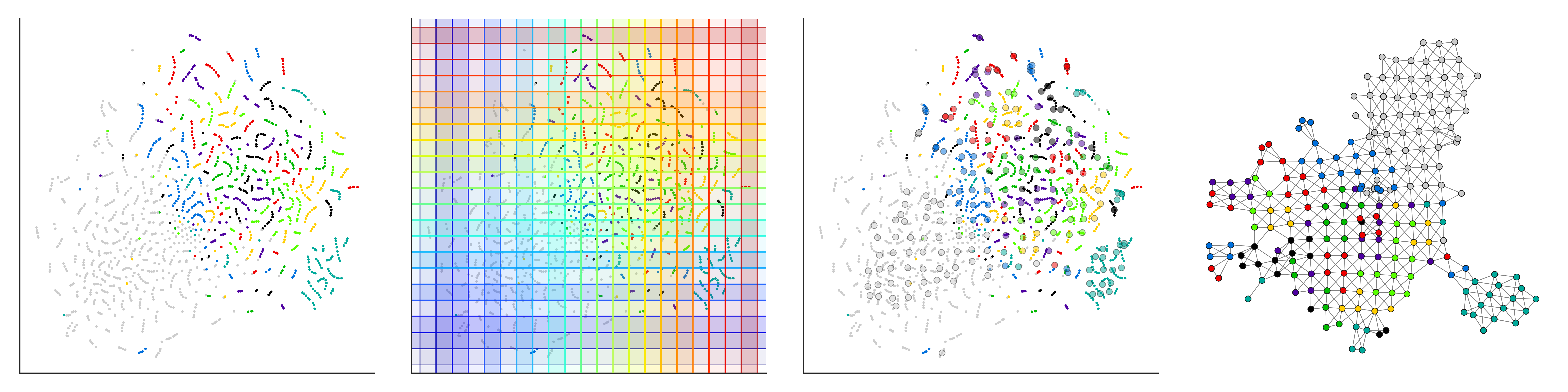
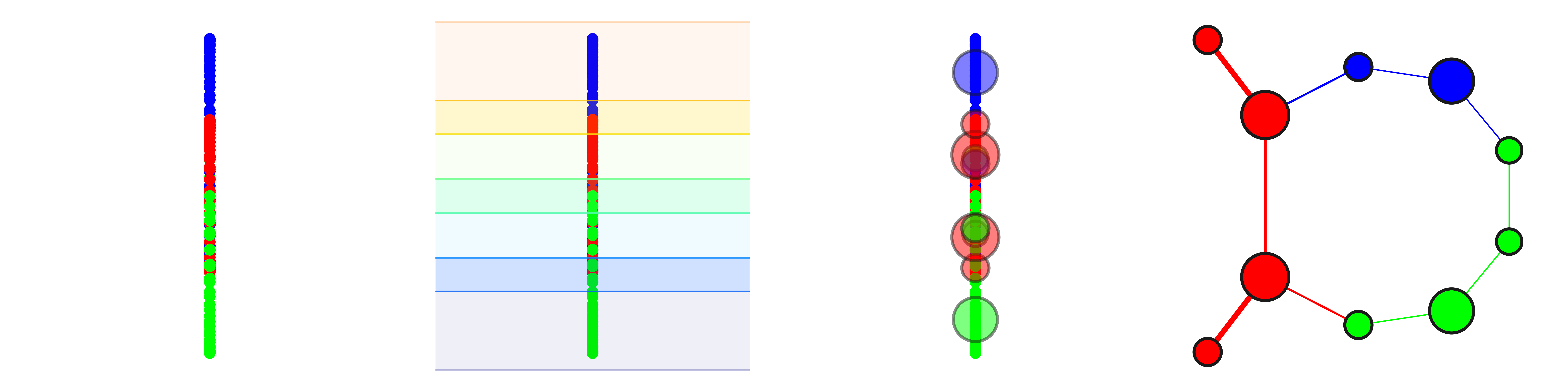
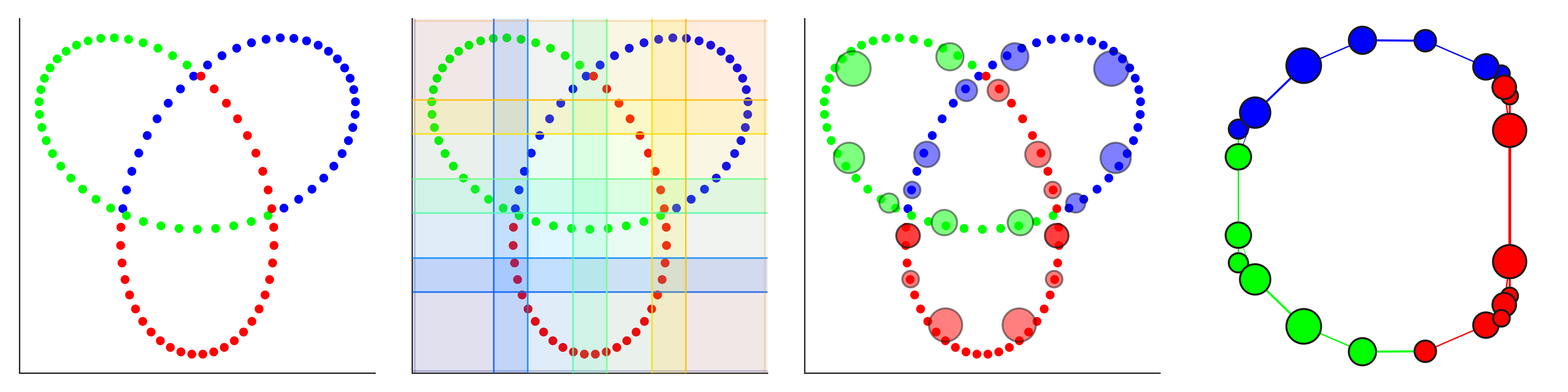
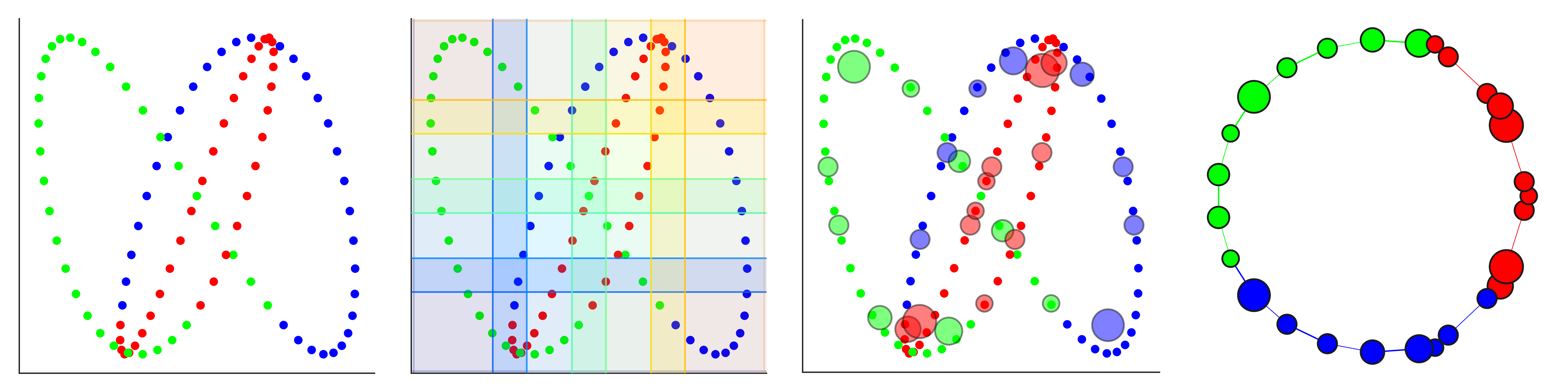
DyNeuSR is a Python visualization library for topological representations of neuroimaging data.
DyNeuSR was designed specifically for working with shape graphs produced by the Mapper algorithm from topological data analysis (TDA), as described in the papers "Generating dynamical neuroimaging spatiotemporal representations (DyNeuSR) using topological data analysis" (Geniesse et al., 2019) and "Towards a new approach to reveal dynamical organization of the brain using topological data analysis" (Saggar et al., 2018). Check out this blog post for more about the initial work that inspired the development of DyNeuSR.
Developed with neuroimaging data analysis in mind, DyNeuSR connects existing implementations of Mapper (e.g. KeplerMapper) with network analysis tools (e.g. NetworkX) and other neuroimaging data visualization libraries (e.g. Nilearn) and provides a high-level interface for interacting with and manipulating shape graph representations of neuroimaging data and relating these representations back to neurophysiology.
DyNeuSR also provides an interactive web interface for visualizing and exploring shape graphs. To see this visual interface in action, check out the demos.
- Trefoil knot (code)
- Trefoil knot (custom layouts) (code)
- Haxby fMRI data (t-SNE lens) (code)
- Haxby fMRI data (UMAP lens) (code)
- Haxby fMRI data (supervised UMAP lens) (code)
- DyNeuSR Fire is a new project that provides a command line interface for DyNeuSR. It wraps
kmapperanddyneusrinto a single pipeline, and uses the Python Fire library to automatically generate a simple command line interface that accepts several important options and allows users to customize this pipeline. For more information about DyNeuSR Fire, check out the docs.
If you find DyNeuSR useful, please consider citing:
Geniesse, C., Sporns, O., Petri, G., & Saggar, M. (2019). Generating dynamical neuroimaging spatiotemporal representations (DyNeuSR) using topological data analysis. Network Neuroscience. Advance publication. doi:10.1162/netn_a_00093
For more information about the Mapper approach, please see:
Saggar, M., Sporns, O., Gonzalez-Castillo, J., Bandettini, P.A., Carlsson, G., Glover, G., & Reiss, A.L. (2018). Towards a new approach to reveal dynamical organization of the brain using topological data analysis. Nature Communications, 9(1). doi:10.1038/s41467-018-03664-4
DyNeuSR provides a Python API for working with and visualizing shape graphs generated by Mapper. This repository includes several examples that introduce DyNeuSR's API and highlight different aspects of analysis with DyNeuSR. For more detailed tutorials, check out dyneusr-notebooks.
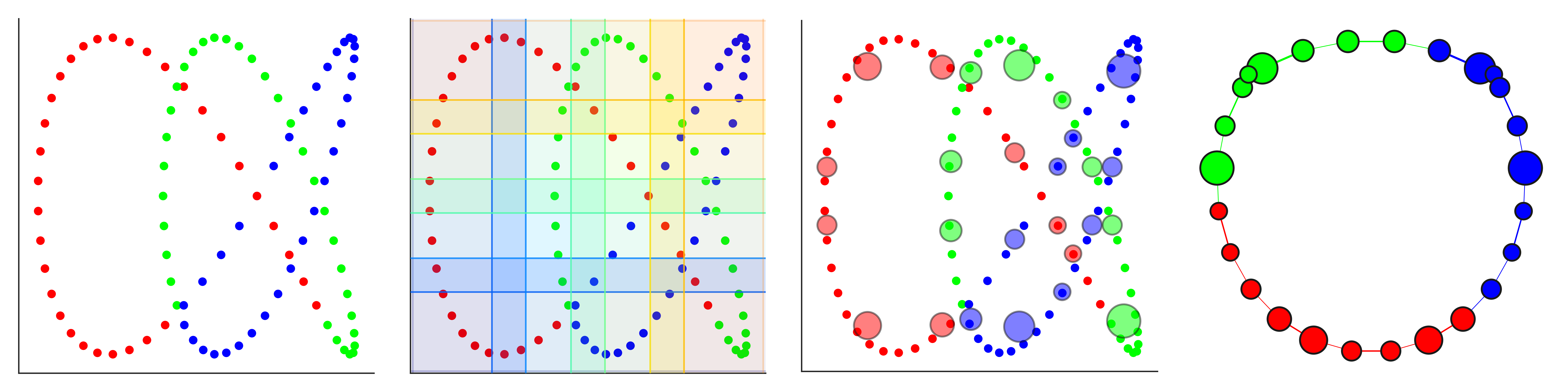
Shape Graph Visualization (trefoil knot)
from dyneusr import DyNeuGraph
from dyneusr.datasets import make_trefoil
from dyneusr.tools import visualize_mapper_stages
from kmapper import KeplerMapper
# Generate synthetic dataset
dataset = make_trefoil(size=100)
X = dataset.data
y = dataset.target
# Generate shape graph using KeplerMapper
mapper = KeplerMapper(verbose=1)
lens = mapper.fit_transform(X, projection=[0])
graph = mapper.map(lens, X, nr_cubes=6, overlap_perc=0.2)
# Visualize the shape graph using DyNeuSR's DyNeuGraph
dG = DyNeuGraph(G=graph, y=y)
dG.visualize('dyneusr_output.html')Mapper Parameter Comparison (trefoil knot)
# Define projections to compare
projections = ([0], [0,1], [1,2], [0, 2])
# Compare different sets of columns as lenses
for projection in projections:
# Generate shape graph using KeplerMapper
mapper = KeplerMapper(verbose=1)
lens = mapper.fit_transform(X, projection=projection)
graph = mapper.map(lens, X, nr_cubes=4, overlap_perc=0.3)
# Visualize the stages of Mapper
fig, axes = visualize_mapper_stages(
dataset, lens=lens, graph=graph, cover=mapper.cover,
layout="spectral")
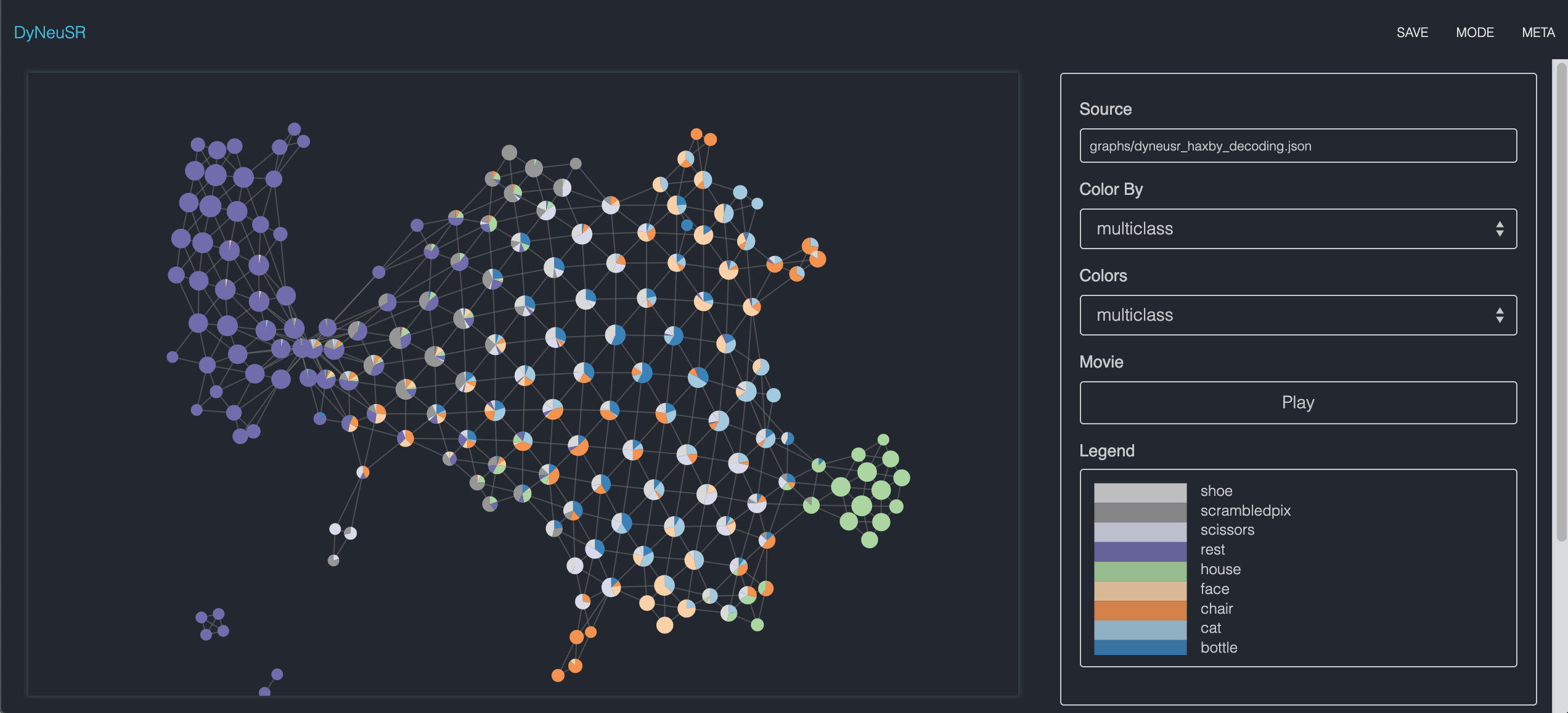
Neuroimaging Applications (haxby decoding)
import numpy as np
import pandas as pd
from nilearn.datasets import fetch_haxby
from nilearn.input_data import NiftiMasker
from kmapper import KeplerMapper, Cover
from sklearn.manifold import TSNE
from sklearn.cluster import DBSCAN
# Fetch dataset, extract time-series from ventral temporal (VT) mask
dataset = fetch_haxby()
masker = NiftiMasker(
dataset.mask_vt[0],
standardize=True, detrend=True, smoothing_fwhm=4.0,
low_pass=0.09, high_pass=0.008, t_r=2.5,
memory="nilearn_cache")
X = masker.fit_transform(dataset.func[0])
# Encode labels as integers
df = pd.read_csv(dataset.session_target[0], sep=" ")
target, labels = pd.factorize(df.labels.values)
y = pd.DataFrame({l:(target==i).astype(int) for i,l in enumerate(labels)})
# Generate shape graph using KeplerMapper
mapper = KeplerMapper(verbose=1)
lens = mapper.fit_transform(X, projection=TSNE(2))
graph = mapper.map(lens, X, cover=Cover(20, 0.5), clusterer=DBSCAN(eps=20.))
# Visualize the shape graph using DyNeuSR's DyNeuGraph
dG = DyNeuGraph(G=graph, y=y)
dG.visualize('dyneusr_output.html')For a full list of packages and required versions, see requirements.txt and requirements-versions.txt.
To install with pip:
pip install dyneusrTo install from source:
git clone https://github.com/braindynamicslab/dyneusr.git
cd dyneusr
pip install -r requirements.txt
pip install -e .
pytestIf your default environment is Python 2, we recommend that you install dyneusr in a separate Python 3 environment. You can find more information about creating a separate environment for Python 3 here.
If you don't have conda, or if you are new to scientific python, we recommend that you download the Anaconda scientific python distribution.
To create a new conda environment and install from source:
conda create -n dyneusr python=3.6
conda activate dyneusr
git clone https://github.com/braindynamicslab/dyneusr.git
cd dyneusr
conda install --file requirements-conda.txt
pip install -e .
pytestThis creates a new conda environment dyneusr and installs in it the dependencies that are needed. To access it, use the conda activate dyneusr command (if your conda version >= 4.4) and use source activate dyneusr command (if your conda version < 4.4).
To run dyneusr in a Singularity container, for use on clusters where you do not have root access for example, first install singularity. Then, you can use the following command:
singularity run shub://braindynamicslab/dyneusr
source activate neuro
This will download and run a singularity container from singularity-hub running centos 7 with dyneusr, jupyter, and all dependencies. It will also activate the conda environement neuro where these are installed. For more information on how to use singularity, see the documentation. The singularity recipe for this file was built with neurodocker
Please feel free to report any issues, request new features, or propose improvements. You can also contact Caleb Geniesse at geniesse [at] stanford [dot] edu.
If you contribute to DyNeuSR, please feel free to add your name to the list of contributors.
Geniesse, C., Sporns, O., Petri, G., & Saggar, M. (2019). Generating dynamical neuroimaging spatiotemporal representations (DyNeuSR) using topological data analysis. Network Neuroscience. Advance publication. doi:10.1162/netn_a_00093