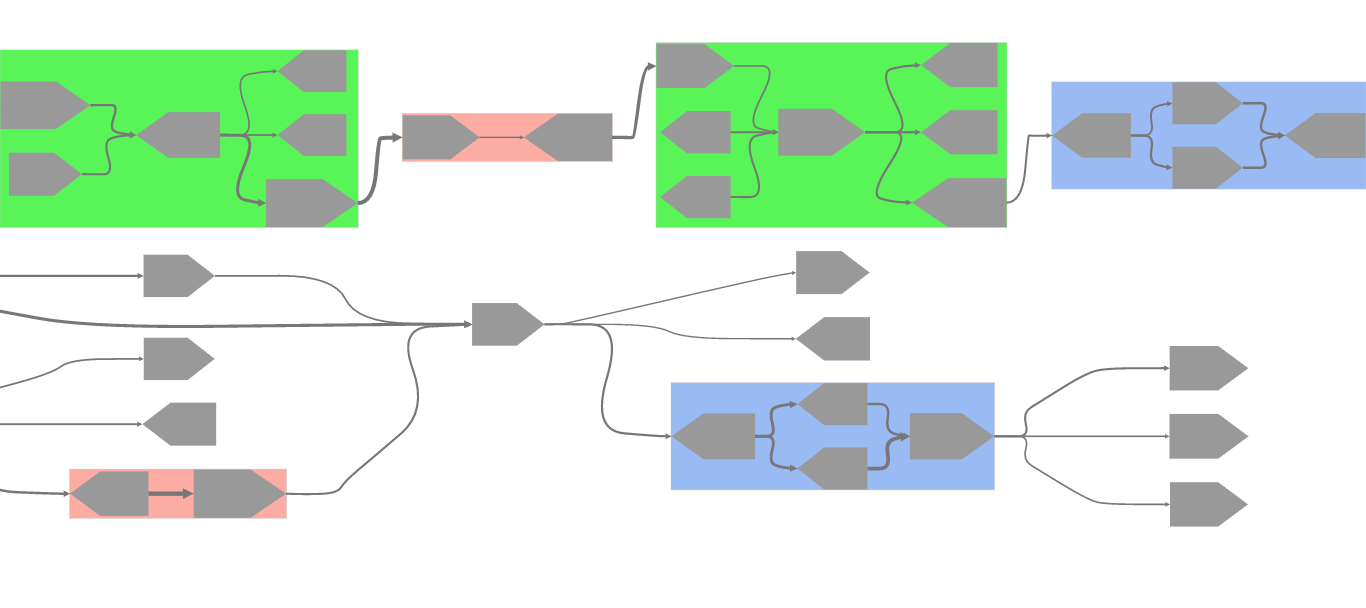
An interactive visualization tool designed for metagenomic sequence assembly and scaffold graphs. The tool aims to display a semi-linearized, hierarchical overview of the input graph while emphasizing the presence of certain structural patterns in the graph.
To this end, MetagenomeScope highlights certain patterns of contigs in the
graph (bubbles, frayed ropes, chains, and "linear" cycles),
splits the graph into its connected components (only displaying one connected
component at a time),
and uses Graphviz' dot tool to hierarchically
lay out each connected component of a graph.
MetagenomeScope also supports the use of
SPQR tree decompositions
(computed using OGDF) to present an
iteratively expandable hierarchical overview of the biconnected components of
the graph.
MetagenomeScope is composed of two main components:
-
The preprocessing script (contained in the
graph_collator/directory of this repository), a Python and C++ script that takes as input an assembly or scaffold graph file and produces a SQLite .db file that can be visualized in the viewer interface.collate.pyis the main script that needs to be run here; it usesspqr.cppto interface with OGDF to generate SPQR tree decompositions. This preprocessing step takes care of graph layout, pattern detection, and SPQR tree generation. Currently, this supports LastGraph (Velvet), GML (MetaCarvel), and GFA input files. Support for GFA2 and FASTG (SPAdes) files is planned. -
The viewer interface (contained in the
viewer/directory of this repository), a client-side web application that reads a .db file generated bycollate.pyand renders the resulting graph using Cytoscape.js. This is coupled with an interface and "control panel" supporting various features to help with assembly finishing and exploratory analysis. (See the section below on using the viewer interface for an explanation of its current features.)
The bifurcated nature of the tool lends it a few advantages that have proved beneficial when analyzing large graphs:
- The user can save a .db file and visualize the contents of the file an arbitrary number of later times, without incurring the costs of layout/pattern detection/etc. twice
- The user can host the viewer interface and a number of .db files on a server, allowing many users to view graphs with the only costs incurred being those of rendering the graphs in question
- 64-bit Linux system (in order to run the C++ binary contained in
graph_collator/spqr) -- however, I'm working on getting a makefile set up to make this platform-independent eventually (see #55) - Python 2.7
- NumPy
- Graphviz, with the
dotandsfdplayout programs installed- Using a version after
2.41.20170712.0019is recommended (see this issue for details).
- Using a version after
- PyGraphviz
- Any modern internet browser (with JavaScript enabled) should be fine. At present, Google Chrome and Mozilla Firefox are recommended, since I haven't done a lot of testing on other browsers yet. (If you run into any problems using the viewer interface on your browser of choice, please let me know and I can look into it.)
collate.py is located in the graph_collator folder. It can be
run from the command line;
see the system requirements section above
for information on what software needs to be installed.
Running collate.py will process an assembly/scaffold graph file so that
it can be visualized. The syntax for this is
./collate.py [-h] -i INPUTFILE -o OUTPUTPREFIX [-d OUTPUTDIRECTORY] [-pg] [-px] [-w] [-b BICOMPONENTSFILE]
The script will always produce a .db file that can be loaded in the viewer
application to visualize the assembly graph.
If the -pg argument is passed, .gv files (in the DOT language)
for each connected component of the assembly graph will be generated; if the
-px argument is passed, .xdot files (in the xdot language) for each
connected component of the assembly graph will be generated.
The script will also generate a few types of auxiliary files containing various information about the structure of the assembly graph. These files are:
prefix_links, whereprefixis the output prefix passed via-o. Only one of these files will be generated per execution ofcollate.py. This file indicates all the edges in the assembly graph. If you pass in-band the input assembly graph has unoriented contigs, then this file will not be generated (since it would be equivalent to the _single_links file in that case).prefix_single_links, whereprefixis the output prefix passed via-o. This file will only be generated if the input assembly graph has unoriented contigs. In terms of currently supported input filetypes, this means that this file will only be generated when the input assembly graph is of type LastGraph or GFA.prefix_bicmps, whereprefixis the output prefix passed via-o. Only one of these files will be generated per execution ofcollate.py. This file indicates the various separation pairs contained within the assembly graph (see Nijkamp et al. for a brief overview of separation pairs and their usage in bubble detection). It's possible to pass an existing version of this file using-bto the script, to prevent having to do the work of creating the file again.component_D.info, whereDis an integer greater than 0. There will be one of these files created for every biconnected component contained within the assembly graph: these files indicate the contents of the SPQR tree defined for their corresponding biconnected component.spqrD.gml, whereDis an integer greater than 0. These files correspond tocomponent_D.infofiles: they indicate the connections between the metanodes of a SPQR tree.
The script requires all component_D.info and spqrD.gml files to be
removed from the output directory before it generates more of them.
If -w is enabled, then all existing files with corresponding names in the
output directory will be deleted; however, if -w is not enabled, then an
error will be raised.
Similarly, if files exist in the output directory with filenames overlapping
those of the prefix_links and prefix_bicmps files, then those files will be
either deleted (if -w is enabled) or an error will be raised (if -w is not
enabled).
-iThe input assembly graph file to be used.-oThe file prefix to be used for all files generated. As an example, given the argument-o prefix, the fileprefix.dbwould be generated. If .gv and/or .xdot files are created (depending on the-pgor-pxarguments, respectively), then those files will be numbered according to the relative size rank (in nodes) of their respective connected component within the assembly graph.-dThis optional argument specifies the name of the directory in which all output files will be stored. If this argument is not indicated, then all files will be generated in the current working directory.-pgThis optional argument produces DOT files (suffix .gv) in the output directory. As an example, given the arguments-o prefixand-pgfor an assembly graph with 3 connected components, the filesprefix.db,prefix_1.gv,prefix_2.gv, andprefix_3.gvwould be created (whereprefix_1.gvindicates the largest connected component by number of nodes,prefix_2.gvindicates the next largest connected component, and so on).-pxThis optional argument produces .xdot files in the output directory. These files are labelled in an identical fashion to.gvfiles, with the only difference in naming being the file suffix (.xdot instead of .gv).-bThis optional argument lets you pass in an existing file indicating the separation pairs in the graph (to be used in the detection of complex bubbles) to the script.-wThis optional argument allows the overwriting of output files (.db/.xdot/.gv/links/single_links/bicmps/.info/spqr.gml files). If this argument is not given, then:- An error will be raised if writing a .db file would cause another .db file to be overwritten.
- A warning will be displayed if writing to a .gv or .xdot file would cause another .gv/.xdot file to be overwritten. In this case, the .gv/.xdot file in question simply would not be saved.
- Note that the presence of files in the
output directory that are conflicting-named folders (e.g. a
directory named
e_coli.db/in the output directory while attempting to produce a file namede_coli.db) will cause an error/warning to be raised regardless of whether or not-wis set.
Graphviz seems to round input node dimensions to the nearest point value (where an inch is defined as 72 points). See this issue for details on the rounding process.
We don't use these rounded dimensions
in the viewer interface, although the rounded dimensions will persist in
.xdot files and when Graphviz performs layout on/draws the .gv files
produced via -pg.
This results in a very slight discrepancy in node sizes between the viewer
interface and Graphviz' drawings.
To save time and space, we don't actually call Graphviz to lay out connected
components containing one node and no edges or node groups. Instead, we
position each node in the center of an appropriately-sized connected component,
essentially "mimicking" the layout PyGraphviz/Graphviz would have produced in a
portion of the time taken to invoke them. (See
this issue for
details.) Therefore, .gv/.xdot files for these connected components will not be
exported even if -pg or -px are passed.
You can use the interface in any modern web browser. Chrome/Firefox are recommended, but most modern browsers should be fine.
A variety of settings are available in the viewer interface. These are
accessible via the Settings button located near the top of the controls
panel, which should be viewable on the left side of the application's display.
There are three categories of settings, each housed in its own tab: animation, performance, and colors.
The viewer interface contains a few features designed to help provide the user with information as certain events occur -- for example, the progress bar is updated as files are loaded, text is displayed as certain types of elements in the graph are drawn, and disabled buttons are displayed as semitransparent.
Some of these ways in which the viewer interface is updated are, or have the potential to be, relatively fast. For example, the process of drawing a small graph might cause the progress bar to rise from 0% to 100% quickly. Or the enabling/disabling of certain buttons (for example, during the process of drawing a graph), and the resulting changes of the buttons' styles, might seem jarring to some users.
A few settings are available that you can use to disable some of the visual updates provided by the viewer interface. Please note, however, that these settings are by no means comprehensive.
- If you would like to turn off the stripe effect used to show an
"indeterminate"
status in the progress bar, you can disable the
Show moving stripes on indeterminate progress barssetting. - If you would like to turn off the text messages displayed underneath the
progress bar as a connected component is drawn, you can disable the
Update status text while drawing connected componentssetting.
The settings here can be used to obtain slight increases in performance of the application when visualizing large connected components. (As with the animation settings, these settings are definitely not comprehensive -- other settings may be added in the future to yield more performance benefits.)
You can change the colors used in various parts of the graph drawings in this settings tab. Configurations of colors can also be exported to a .tsv file; these files can later be imported into the viewer interface, to avoid having to manually set the color settings multiple times.
Database files generated by the preprocessing script that are stored on your device locally can be loaded in the viewer interface using the "Choose .db file" button. In the demo of MetagenomeScope, a number of sample database files are available for you to try out using the "Demo .db" button.
Once you've loaded a file, the "Draw connected component" buttons can be used to render a given connected component in the assembly/scaffold graph represented by the database file in question. Note that connected components are automatically sorted in descending order of number of nodes -- so connected component 1 will be the largest, followed by 2, etc. In assembly graphs, the number of connected components in the "standard mode" view of the graph may be different from the number of connected components in the "decomposition mode" view of the graph -- see the "Assembly info" button for information on the number of connected components in each mode.
This mode draws a normal view of the input graph, rendering each contig as either one (if the input was a scaffold graph) or two (if the input was an assembly graph) nodes. Edges between contigs are drawn accordingly.
In standard mode, structural patterns contained within the graph were automatically highlighted and grouped together during layout. These patterns can be easily identified by their background color and general structure.
If the input assembly graph file has edge frequencies given (either as bsize
in GML files or as a multiplicity attribute in LastGraph files), then edges
in every connected component will be relatively scaled by their frequency.
That is, edges with relatively
higher frequency values than other edges in their connected component of the
graph will be made thicker than other edges in the viewer interface, and
edges with relatively low frequency values will be made thinner.
To account for "outlier" edges (which might be far more frequent or far less frequent than other edges in a connected component, and therefore skew the relative scaling process), we use Tukey fences to detect high and low outlier edge frequencies for each connected component. High outlier edges are given the maximum edge thickness and colored red, and low outlier edges are given the minimum edge thickness and colored blue. (These color assignments can be modified in the color settings, though.) The remaining non-outlier edges in a connected component are scaled relatively, without taking detected outlier edges' frequencies into account.
You can use the View Scaffolds section of the control panel to visualize
scaffolds described within an
AGP file.
Clicking on a scaffold in this panel will select all the nodes contained within
it.
It's assumed that AGP files loaded for assemblies that were originally in the
GML format (i.e. MetaCarvel output) use node labels for the component_id
fields.
It's also assumed that AGP files loaded for other types of assemblies use
node IDs for the component_id fields.
In either case, the ID of a node group in standard mode can additionally be
used as a component_id in AGP files to refer to that node group as a part of
a scaffold.
The buttons in the Assembly Finishing section of the control panel can be
used to select paths of contigs in the graph and export the resulting node
labels (if exporting an AGP file and the current .db file originated from
GML input) or IDs (if not exporting an AGP file, or if the current .db file
originated from other input filetypes).
Currently, paths can be exported as either a comma-separated (CSV) list of node IDs, or as a scaffold in the AGP file format.
In these modes, each biconnected component within the input graph is collapsed into the root node of its corresponding SPQR tree. These trees can then be iteratively expanded via right-clicking (similar to how node groups are collapsible/uncollapsible in standard mode).
In the "implicit" decomposition mode, expansions to the root node of a SPQR tree show iteratively larger amounts of detail added to the root. In the "explicit" decomposition mode, expansions actually result in the immediate descendant metanodes in the SPQR tree being drawn, revealing more and more of the literal SPQR tree structure for a given biconnected component.
Although "implicit" and "explicit" decompositions both show the same amount of nodes and edges upon first being drawn, the layouts involving explicit decomposition will usually be messier (since more space needs to be allocated for the SPQR tree structures) and will involve more nodes than their corresponding implicit decomposition upon fully being uncollapsed.
MetagenomeScope is licensed under the GNU GPL, version 3.
Feel free to let me know if you have any suggestions, comments, or other feedback about the tool.
I can be reached via email at mfedarko at umd dot edu.
You can also open an issue in this repository, if you'd like.
- The preprocessing script (in
collate.py) uses Graphviz'dotandsfdplayout programs via PyGraphviz. - The preprocessing script (in
collate.py) also uses NumPy to calculate percentiles during edge thickness scaling. - The preprocessing script (in
spqr.cpp) uses OGDF to construct SPQR trees.spqr.cppalso uses cmdline.h to parse command-line arguments.
- Both the preprocessing script and the viewer interface use sqlite3. In particular, the preprocessing script uses the built-in Python sqlite3 module while the viewer interface uses sql.js.
- The viewer interface uses Cytoscape.js to render
graphs on the client side.
- Also, the toggling protocol used for the control panel of the viewer interface was inspired by a similar mechanism used in this Cytoscape.js demo.
- The viewer interface uses jQuery and
Bootstrap for various stylistic and functional
purposes in the application.
- The icons used to theme various controls in the viewer application are from the Glyphicon Halflings set, included with Bootstrap.
- The color selection functionality in the viewer interface uses the Bootstrap Colorpicker plugin.