This SQLite3 loadable extension adds features to the ubiquitous embedded RDBMS supporting applications in genome bioinformatics:
- genomic range indexing for overlap queries & joins
- in-SQL utility functions, e.g. reverse-complement DNA, parse "chr1:2,345-6,789"
- automatic streaming storage compression (also available standalone)
- reading directly from HTTP(S) URLs (also available standalone)
- pre-tuned settings for "big data"
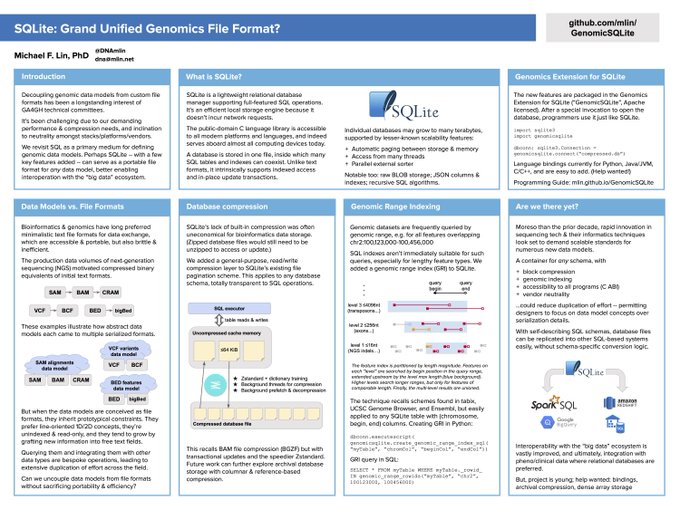
This November 2021 poster discusses the context and long-run ambitions:
Our Colab notebook demonstrates key features with Python, one of several language bindings.
USE AT YOUR OWN RISK: This project is not associated with the SQLite developers. The database storage extensions are designed to preserve ACID transaction safety, but they're young and unlikely to be totally bug-free.
Start Here 👉 full documentation site
We supply the extension prepackaged for Linux x86-64 and macOS Catalina. An up-to-date version of SQLite itself is also required, as specified in the docs.
Programming language support:
- C/C++
- Python ≥3.6
- Java & JVM languages
- Rust
More to come. (Help wanted; see Language Bindings Guide)
Most will prefer to install a pre-built shared library (see above). To build from source, see our Actions yml (Ubuntu 20.04) or Dockerfile (CentOS 7) used to build the more-portable releases. Briefly, you'll need:
- C++11 build system
- CMake ≥ 3.14
- Dev packages: SQLite ≥ 3.31.0, Zstandard ≥ 1.3.4, libcurl
And incantations:
cmake -DCMAKE_BUILD_TYPE=Release -B build .
cmake --build build -j 4 --target genomicsqlite
...generating build/libgenomicsqlite.so. To run the test suite, you'll furthermore need:
- htslib ≥ 1.9, samtools, and tabix
- pigz
- Python ≥ 3.6 and packages: pytest pytest-xdist pre-commit black pylint flake8
- JDK, mvn, rust
- clang-format & cppcheck
to:
pre-commit run --all-files # formatters+linters
cmake -DCMAKE_BUILD_TYPE=Debug -B build .
cmake --build build -j 4
env -C build ctest -V