-
Full integration with
aptas every binary resolves all its dependencies: No more installations (of pre-built archives) only to discover that a shared library is missing. No more surprises. -
Full integration with
aptso that an update of a system library cannot break an R package: if a (shared) library is used by a CRAN, the package manager knows, and will not remove it. No more (R package) breakage from (system) library updates. -
Simpler and lighter than some alternatives as only run-time library packages are installed as dependencies (instead of generally heavier development packages).
-
Installations are fast, automated and reversible thanks to the package management layer.
-
Fast and well-connected mirror at r2u.stat.illinois.edu on the Internet2
-
Complete coverage with (currently) ~ 22,651 CRAN packages (and 401 from BioConductor) using current versions: We use R 4.3.2, and BioConductor 3.18.
-
Complete support for both Ubuntu 20.04 ("focal") and Ubuntu 22.04 ("jammy").
-
Optional (but recommended) bspm use automagically connects R functions like
install.packages()toaptfor access to binaries and dependencies. -
Docker containers
rocker/r2ufrom the Rocker Project for both 'focal' and 'jammy'. -
GitHub Actions support to set up on Ubuntu 22.04 "jammy" or via container
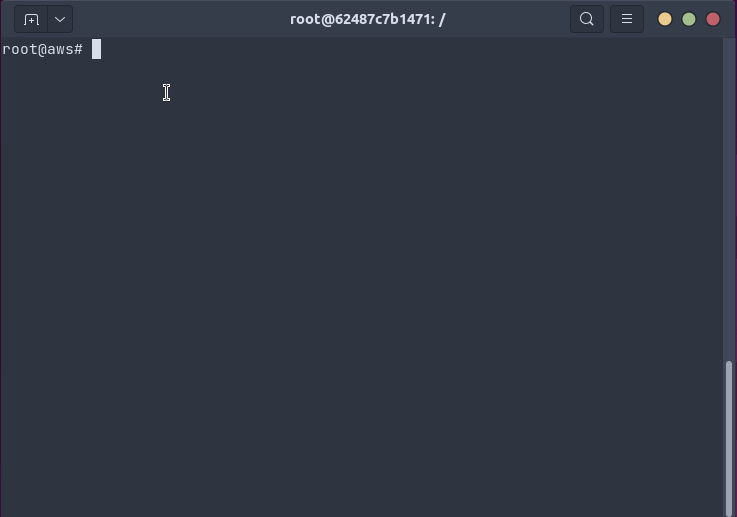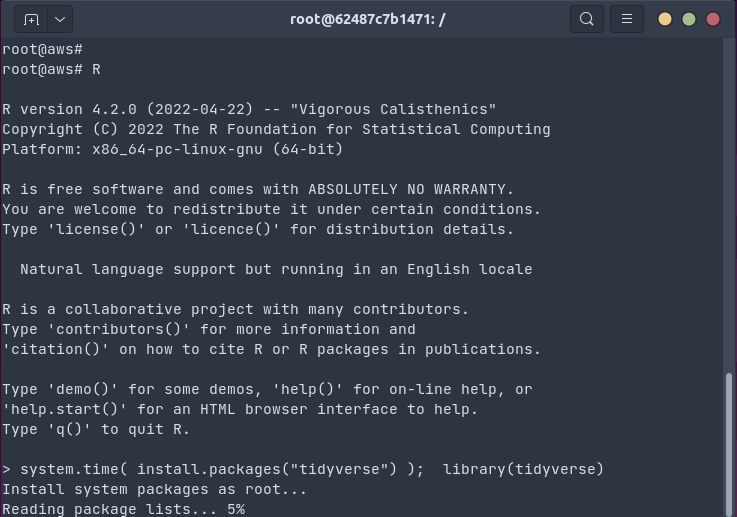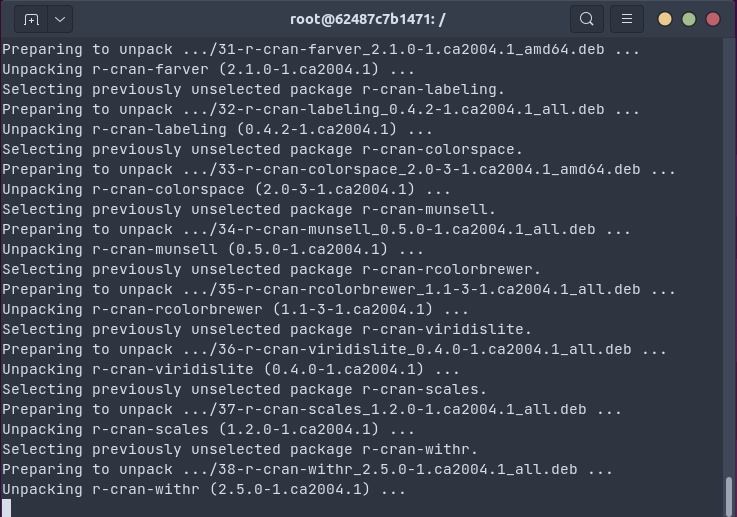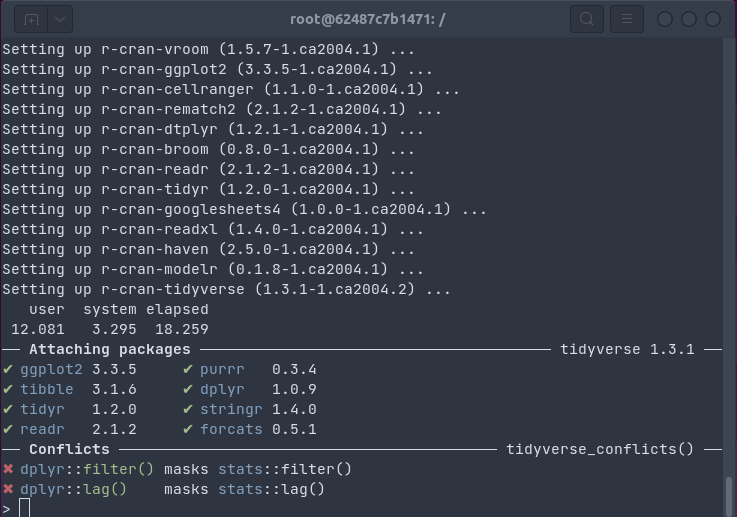
The gif below shows how one install.packages("tidyverse") command on an Ubuntu
20.04 system installs all packages and dependencies as binaries in 18 seconds (by passing the
R package installation to apt using bspm).
This uses the Docker container referenced below, which has been set up with the five easy setup steps detailed here.
We generally support amd64 (i.e. standard 64-bit Intel/AMD cpus, sometimes also called x86_64) for the current Ubuntu LTS release and its predecessor release (more on this here). We use 'r-release' just like CRAN. So currently the 'focal' 20.04 LTS and 'jammy' 22.04 LTS releases are fully supported.
Support for other cpu architectures is certainly possible but somewhat unlikely due to a lack of (additional hardware) resources and time. Support for other distributions is possible but unlikely right now (due to a lack of resources and time). P3M/PPM/RSPM now appears to also support Debian which could be added at some later point.
Current versions are based on R 4.3.2, and BioConductor release 3.18 packages are provided when required by CRAN packages. Binaries are generally R 4.3.* based. Some older packages released when we used R 4.2.* may have been built with R 4.2., they will still work the same with R 4.3. as R is generally forward-compatible.
Everything :)
Initially, we started from cran-logs by picking the N most-downloaded packages, along with their dependencies from BioConductor. (It should be noted that for example the first 100 packages already account for approximately half the total downloads: it is a very skewed distribution.) We iterated, and fairly soon arrived of full coverage of CRAN.
So we now cover
- all CRAN packages (modulo at best handful of blacklisted ones) including all packages needing compilation
- all BioConductor packages implied by these plus a 'healthy subset' of the highest scoring BioConductor packages (also covering e.g. all BioConductor packages in the Debian and Ubuntu distributions)
This currently results in 22741 and 22651 binary packages from CRAN in "focal" and "jammy", respectively, and 401 and 401 BioConductor packages, respectively, from the 3.18 release. (See this FAQ about why this number is higher than CRAN.)
The sole exception are two packages we cannot build (as we do not have the required commercial software it accessess) plus less than a handful of 'odd builds' that fail and are skipped.
For the CRAN binaries we either repackage RSPM/PPM builds (where available) or build natively. All selected BioConductor 3.18 packages are built natively. For all of these, full dependency resolution and integration with the system is a key feature.
Everything is provided as .deb binary files with proper dependency
resolution by using a proper apt repo which also has a signed Release file.
(Note that you could use one of the two scripts
add_cranapt_jammy.sh
(for Ubuntu 22.04), or
add_cranapt_focal.sh
(for the older Ubuntu 20.04) to facilitate the setup. They are tested on 'empty' Ubuntu containers
of the corresponding release. However, you may prefer to execute the steps outlined here by hand.)
We show the setup step by step for 'jammy' aka Ubuntu 22.04. You should run all these commands
as root to carefully review each one.
First add the repository key so that apt knows it (this is optional but recommended)
apt update -qq && apt install --yes --no-install-recommends wget \
ca-certificates gnupg
wget -q -O- https://eddelbuettel.github.io/r2u/assets/dirk_eddelbuettel_key.asc \
| tee -a /etc/apt/trusted.gpg.d/cranapt_key.ascSecond, add the repository to the apt registry. We recommend the well-connected main mirror
provide at University of Illinois:
echo "deb [arch=amd64] https://r2u.stat.illinois.edu/ubuntu jammy main" \
> /etc/apt/sources.list.d/cranapt.list
apt update -qqThird, and optionally, if you do not yet have the current R version, run these two lines (or use the standard CRAN repo setup)
wget -q -O- https://cloud.r-project.org/bin/linux/ubuntu/marutter_pubkey.asc \
| tee -a /etc/apt/trusted.gpg.d/cran_ubuntu_key.asc
echo "deb [arch=amd64] https://cloud.r-project.org/bin/linux/ubuntu jammy-cran40/" \
> /etc/apt/sources.list.d/cran_r.list
apt-key adv --keyserver keyserver.ubuntu.com --recv-keys \
67C2D66C4B1D4339 51716619E084DAB9
apt update -qq
DEBIAN_FRONTEND=noninteractive apt install --yes --no-install-recommends \
r-base-coreFourth, add repository 'pinning' as apt might get confused by some older
packages (in the Ubuntu distro) which accidentally appear with a higher
version number. See the next section for a short discussion how it ensures 'CRANapt'
sorts highest.
echo "Package: *" > /etc/apt/preferences.d/99cranapt
echo "Pin: release o=CRAN-Apt Project" >> /etc/apt/preferences.d/99cranapt
echo "Pin: release l=CRAN-Apt Packages" >> /etc/apt/preferences.d/99cranapt
echo "Pin-Priority: 700" >> /etc/apt/preferences.d/99cranaptAfter that the package are known (under their r-cran-* and r-bioc-*
names). You can install them on the command-line using apt and apt-get,
via aptitude as well as other front-ends.
Fifth, and also optional, install and enable the bspm
package so that the r2u (or CRANapt) as well as other R packages (available as r-*.deb binaries)
become available via install.packages() and update.packages(). Note that you may need to install
it directly from source via sudo Rscript -e 'install.packages("bspm")' to ensure it integrates
correctly with the packaging system. You should also install Python components used internally by
bspm via the sudo apt-get install python3-{dbus,gi,apt} command.
apt install --yes --no-install-recommends python3-{dbus,gi,apt}
## Then install bspm (as root) and enable it, and enable a speed optimization
Rscript -e 'install.packages("bspm")'
RHOME=$(R RHOME)
echo "suppressMessages(bspm::enable())" >> ${RHOME}/etc/Rprofile.site
echo "options(bspm.version.check=FALSE)" >> ${RHOME}/etc/Rprofile.siteThat's it! Now try it out!
Packages can be found in different repositories, and generally the highest available version is
the one we what---and apt picks it for us. Now, because we let apt (and related tools) pick the
packages based on versions, we may want to ensure that the CRANapt repo sorts higher than the
default repo as (older) package builds in the distribution itself may appear (to apt) to be newer
via a quirk in the sorting algorithm. A case in point was package gtable whose version in Ubuntu
was 0.3.0+dfsg-1 which accidentally sorts higher than the rebuild we made under a newer and more
consistent version number 0.3.0-1.ca2004.1.
For this issue, one possible and popular fix is to use 'apt pinning'. It can give 'higher weight' to packages from a particular repositor or tag. In the suggested example above, we give the r2u / cranapt repo a weight of 700 which is higher than the package default value of 500.
There are also two Docker containers for Ubuntu 20.04 'focal' and 22.04 'jammy', respectively. Initially published as eddelbuettel/r2u, these are now also available also as rocker/r2u. They all have the features detailed above, including pinning and bspm support, already set up.
Each of the Ubuntu LTS flavors, i.e., 'focal' and 'jammy' is also available as an identical image using the release version, i.e., '20.04' and '22.04', respectively.
Note that with some builds of Docker (and possibly related to Ubuntu hosts) you may have to add
the --security-opt seccomp=unconfined option to your Docker invocation to take advantage of bspm
and the full system integration inside the container.
This is also documented in the FAQ.
There are two basic ways to take advantage of r2u in a GitHub Actions. The first, and simplest,
is to switch to using the Docker container (see previous section). This is as simple as adding the
container: statement after runs-on: in jobs: section:
runs-on: ubuntu-latest
container:
image: rocker/r2u:22.04
A complete example is provided in this R package repo. The key advantage of this approach is that everything is already set up.
A second approach consists of adding r2u as a step via the r2u-setup GitHub
Action:
- name: Setup r2u
uses: eddelbuettel/github-actions/r2u-setup@master
A complete example is provided in this
repo
where we use it because using the Docker container approach makes committing back via git a little
harder.
See the vignette Codespaces about how to launch a 'Codespace' directly in your browser, launched from the gitrepo within minutes.
This also works from your vscode installation as a remote codespace.
The vignette has more details.
Use this link below (after possibly signing up for gitpod.io first)
and run one of the three example scripts, or just start R in the terminal window.
The gif below display running one such example to install brms from binaries in a few seconds. Using this requires only (free) GitHub and GitPod accounts.
Usage is vibrant. As of late 2023, nearly 300,000 packages are shipped per week, with a total of now over thirteen million packages shipped. Early September 2023 also had the most recent and dramatic spike of over three million packages in two days. The following chart gives a summary of cumulative and average weekly downloads (the latter one on a log scale) as of December 2023.
Please file issues at the GitHub issues for r2u.
Please also see the FAQ for answers to Frequently Asked Questions.
-
The littler package reflects build-time configuration, the RSPM/PPM binary is then expecting a different R location so it needs a binary rebuild. Added a 'force' flag, may need a list similar to the blacklist to always compiled.
-
A small number of packages do not build for lack required components; examples are ROracle and Rcplex. They, and their reverse dependencies, are blacklisted and not built.
-
r2u is an
aptrepo, which viabspmbecomes used "automagically" via standard R calls ofinstall.packages()and alike. That last part is important: package installations that do not useinstall.packages()(such asrenv,rig, ...) do not benefit frominstall.packages()callingaptfor you, and cannot take advantage of r2u viabspm. -
bspmtraces calls toinstall.packages()and maps them system-wide installation viaapt. By choice, it does not map theremove.packages()for package removal, see this issue for more discussion. Packages can be uninstalled via the system package manager using, respectively,apt,dpkgor one of graphical frontends as well as via the R functionbspm::remove_sys().
Dirk Eddelbuettel
The repository-building code in this package is released under the GPL (>= 2).
All CRAN and BioConductor packages are released under their respective licenses.
This was made possible by the generous support of endless coffee thanks to my GitHub Sponsors.