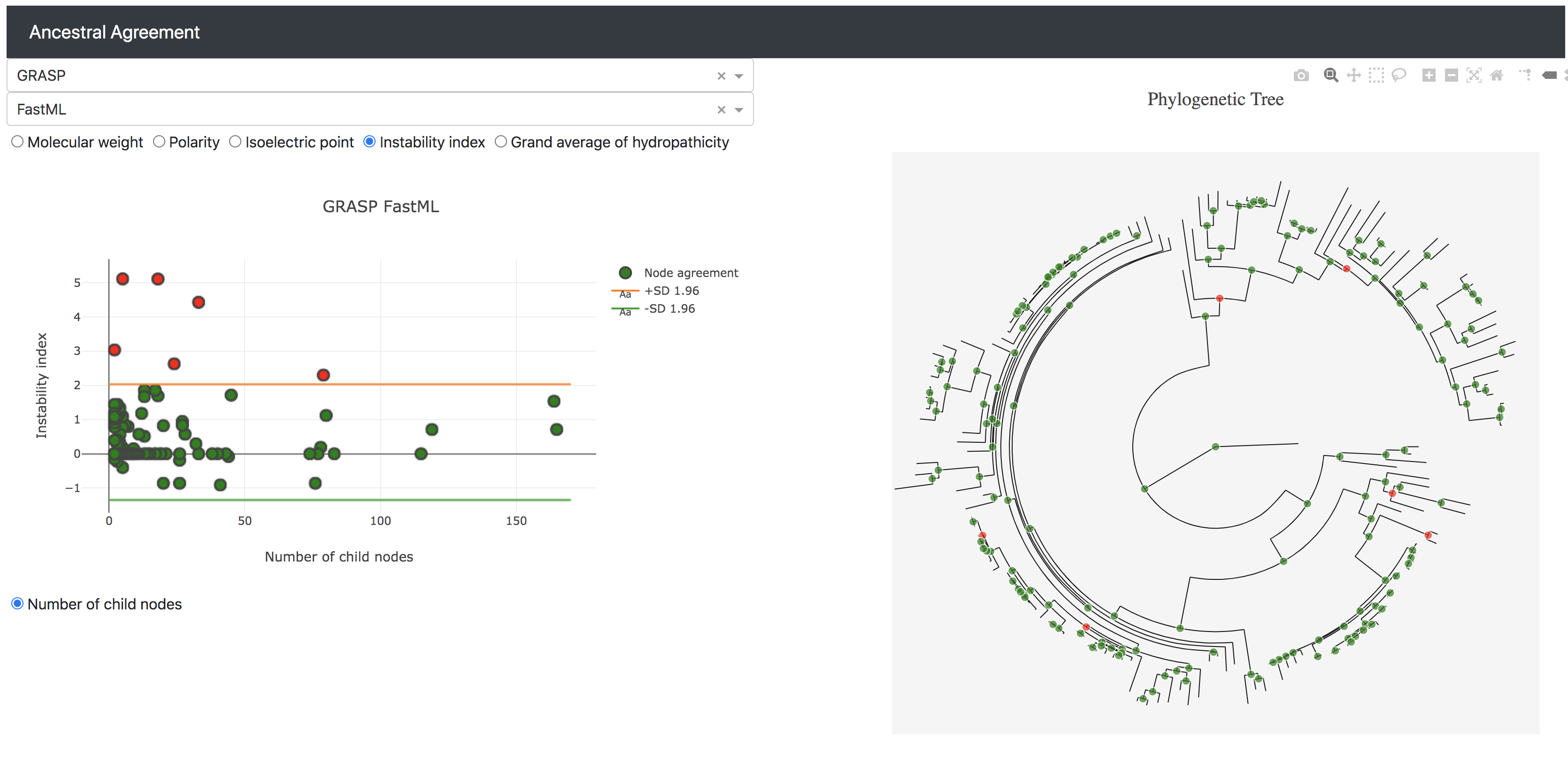
Ancestral Agreement is a method, inspired by Bland-Altman plots, for comparing the agreement between two sets of predicted biological ancestral sequences.
Each pair of ancestral protein sequences between two sets are converted into quantitative predictions for a range of metrics and the differences between sets are plotted against the evolutionary age of each ancestor. Results are easily interpretable and highlight whether particular methods consistently disagree, whether specific nodes are inconsistently inferred, and specific evolutionary times beyond which methods start to diverge.
- Install MongoDB
https://docs.mongodb.com/manual/installation/
You can set the name of the database you want to create in aa/configs/mongoconfig.py and then when you run the application it will create the database for you.
- Clone the repository
git clone github.com/gabe-foley-thesis/ancestral_agreement
cd ancestral_agreement
- Install requirements
Recommended to do this within a conda or virtual environment
pip install -r requirements.txt
python aa/routes.py
You can now open a web browser and go to http://localhost:5000 -
This gives you the upload form which you can enter your data into.
Inputs required are a phylogenetic tree labelled with ancestral node positions (Newick format) and two sets of ancestor sequences from different prediction methods / settings that correspond to the labelled ancestral node positions (FASTA format).
See /files/test_6_1.aln, test_6_2.aln, and test_6.nwk for the expected format.
There is an example data set for a CYP2U1 ancestral reconstruction using either GRASP or FastML. It gets loaded automatically into the Mongo database when you run the app.
If you just want to view the example data then navigate to http://localhost:5000/dash and it will load automatically. Just select the two data sets from the dropdown menus.
After you're loaded the files you need to select the data sets you're interested in viewing from the drop down menu.
Currently the phylogenetic tree must be identical for each method.