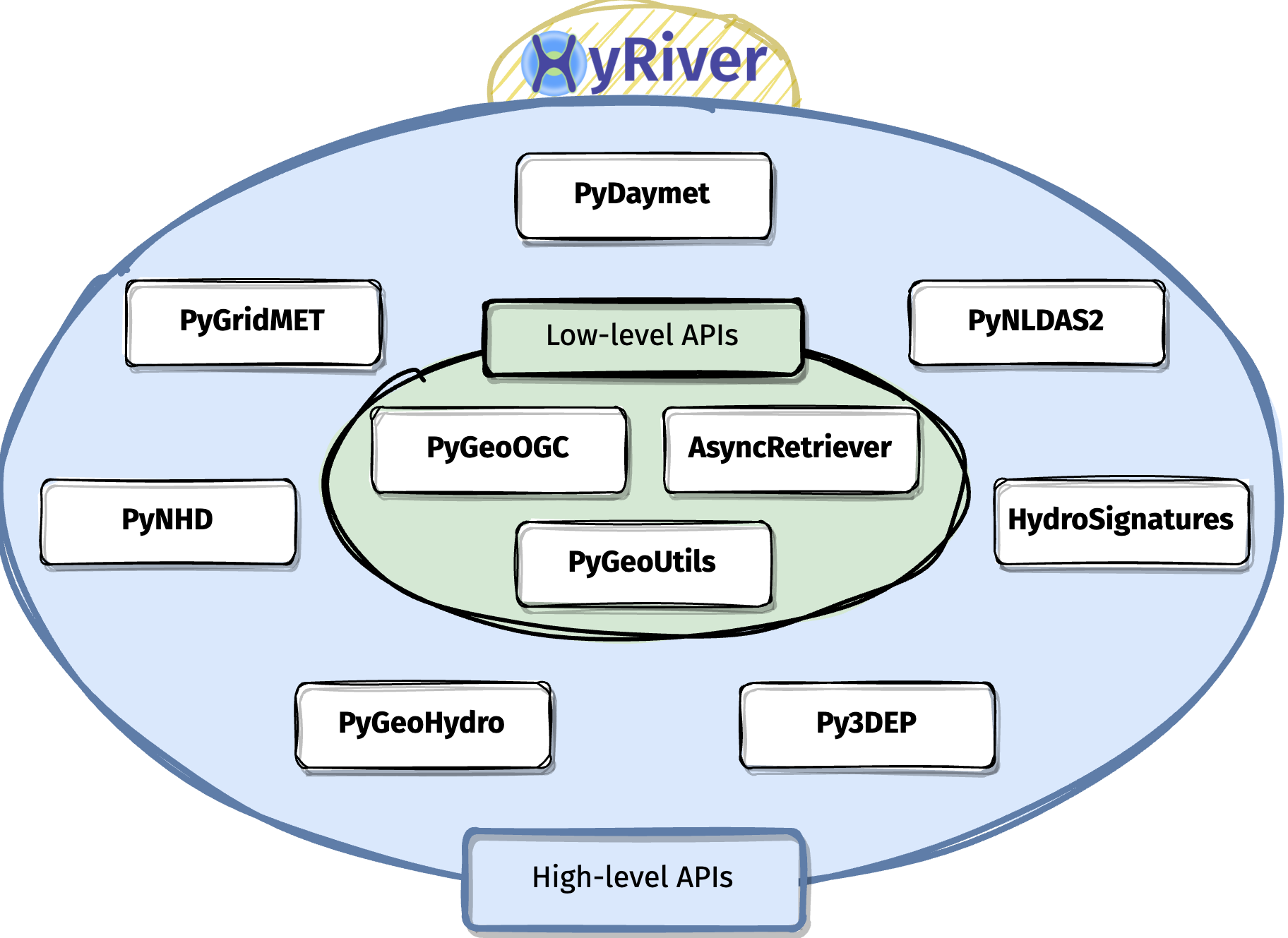
HyRiver is a software stack consisting of ten Python libraries that are designed to aid in hydroclimate analysis through web services. Currently, this project only includes hydrology and climatology data within the US. Some major capabilities of HyRiver are:
- Easy access to many web services for subsetting data on server-side and returning the requests as masked Datasets or GeoDataFrames.
- Splitting large requests into smaller chunks, under-the-hood, since web services often limit the number of features per request. So the only bottleneck for subsetting the data is your local machine memory.
- Navigating and subsetting NHDPlus database (both medium- and high-resolution) using web services.
- Cleaning up the vector NHDPlus data, fixing some common issues, and computing vector-based accumulation through a river network.
- A URL inventory for many popular (and tested) web services.
- Some utilities for manipulating the obtained data and their visualization.
Please visit examples webpage to see some example notebooks. You can also watch these videos for a quick overview of HyRiver capabilities:
You can also try this project without installing it on your system by clicking on the binder badge. A Jupyter Lab instance with the HyRiver software stack pre-installed will be launched in your web browser, and you can start coding!
Please note that this project is in early development stages, while the provided functionalities should be stable, changes in APIs are possible in new releases. But we appreciate it if you give this project a try and provide feedback. Contributions are most welcome.
Moreover, requests for additional databases and functionalities can be submitted via issue trackers of packages.
If you use any of HyRiver packages in your research, we appreciate citations:
@article{Chegini_2021,
author = {Chegini, Taher and Li, Hong-Yi and Leung, L. Ruby},
doi = {10.21105/joss.03175},
journal = {Journal of Open Source Software},
month = {10},
number = {66},
pages = {1--3},
title = {{HyRiver: Hydroclimate Data Retriever}},
volume = {6},
year = {2021}
}You can install all the packages using pip:
$ pip install py3dep pynhd pygeohydro pydaymet pygridmet pynldas2 hydrosignatures pygeoogc pygeoutils async-retrieverPlease note that installation with pip fails if libgdal is not installed on your system. You should install this package manually beforehand. For example, on Ubuntu-based distros the required package is libgdal-dev. If this package is installed on your system you should be able to run gdal-config --version successfully.
Alternatively, you can install them using conda:
$ conda install -c conda-forge py3dep pynhd pygeohydro pydaymet pygridmet pynldas2 hydrosignatures pygeoogc pygeoutils async-retrieveror mambaforge (recommended):
$ mamba install py3dep pynhd pygeohydro pydaymet pygridmet pynldas2 hydrosignatures pygeoogc pygeoutils async-retrieverAdditionally, you can create a new environment, named hyriver with all the packages and optional dependencies installed with mambaforge using the provided environment.yml file:
$ mamba env create -f ./environment.yml