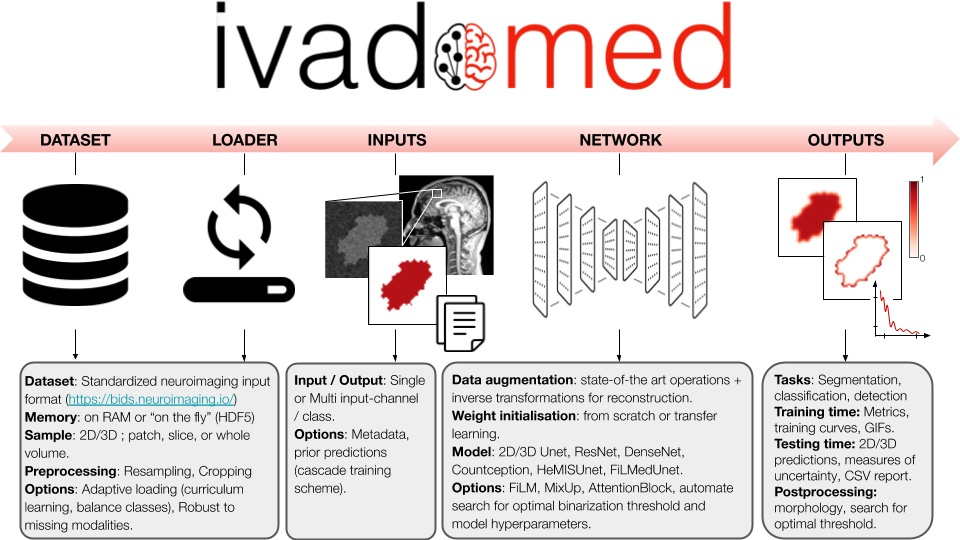
ivadomed is an integrated framework for medical image analysis with deep learning.
The technical documentation is available here. The more detailed installation instruction is available there
ivadomed requires Python >= 3.7 and < 3.10 as well as PyTorch == 1.8. We recommend working under a virtual environment, which could be set as follows:
python -m venv ivadomed_env
source ivadomed_env/bin/activateInstall ivadomed and its requirements from Pypi <https://pypi.org/project/ivadomed/>__:
pip install --upgrade pip
pip install ivadomedBleeding-edge developments builds are available on the project's master branch on Github. Installation procedure is the following:
git clone https://github.com/neuropoly/ivadomed.git
cd ivadomed
pip install -e .This project results from a collaboration between the NeuroPoly Lab and the Mila. The main funding source is IVADO.