Important
Development has begun on Splink 4 on the splink4_dev branch. Splink 3 is in maintenance mode and we are no longer accepting new features. We welcome contributions to Splink 4. Read more on our latest blog.
Splink is a Python package for probabilistic record linkage (entity resolution) that allows you to deduplicate and link records from datasets that lack unique identifiers.
⚡ Speed: Capable of linking a million records on a laptop in around a minute.
🎯 Accuracy: Support for term frequency adjustments and user-defined fuzzy matching logic.
🌐 Scalability: Execute linkage in Python (using DuckDB) or big-data backends like AWS Athena or Spark for 100+ million records.
🎓 Unsupervised Learning: No training data is required for model training.
📊 Interactive Outputs: A suite of interactive visualisations help users understand their model and diagnose problems.
Splink's linkage algorithm is based on Fellegi-Sunter's model of record linkage, with various customisations to improve accuracy.
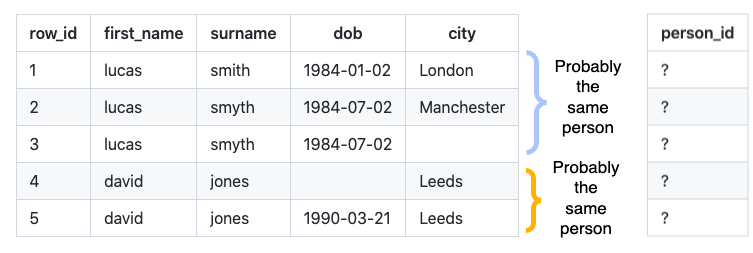
Consider the following records that lack a unique person identifier:
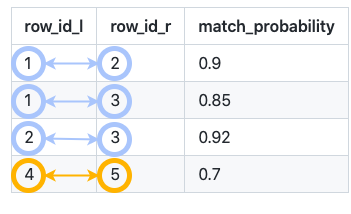
Splink predicts which rows link together:
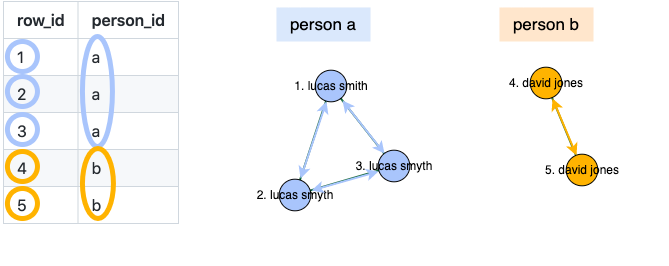
and clusters these links to produce an estimated person ID:
Before using Splink, input data should be standardised, with consistent column names and formatting (e.g., lowercased, punctuation cleaned up, etc.).
Splink performs best with input data containing multiple columns that are not highly correlated. For instance, if the entity type is persons, you may have columns for full name, date of birth, and city. If the entity type is companies, you could have columns for name, turnover, sector, and telephone number.
High correlation occurs when the value of a column is highly constrained (predictable) from the value of another column. For example, a 'city' field is almost perfectly correlated with 'postcode'. Gender is highly correlated with 'first name'. Correlation is particularly problematic if all of your input columns are highly correlated.
Splink is not designed for linking a single column containing a 'bag of words'. For example, a table with a single 'company name' column, and no other details.
The homepage for the Splink documentation can be found here. Interactive demos can be found here, or by clicking the following Binder link:
The specification of the Fellegi Sunter statistical model behind splink is similar as that used in the R fastLink package. Accompanying the fastLink package is an academic paper that describes this model. The Splink documentation site and a series of interactive articles also explores the theory behind Splink.
The Office for National Statistics have written a case study about using Splink to link 2021 Census data to itself.
Splink supports python 3.8+. To obtain the latest released version of splink you can install from PyPI using pip:
pip install splinkor, if you prefer, you can instead install splink using conda:
conda install -c conda-forge splinkFor projects requiring specific backends, Splink offers optional installations for Spark, Athena, and PostgreSQL. These can be installed by appending the backend name in brackets to the pip install command:
pip install 'splink[{backend}]'Should you require a version of Splink without DuckDB, see our section on DuckDBLess Splink Installation.
Click here for backend-specific installation commands
pip install 'splink[spark]'pip install 'splink[athena]'pip install 'splink[postgres]'The following code demonstrates how to estimate the parameters of a deduplication model, use it to identify duplicate records, and then use clustering to generate an estimated unique person ID.
For more detailed tutorial, please see here.
from splink.duckdb.linker import DuckDBLinker
import splink.duckdb.comparison_library as cl
import splink.duckdb.comparison_template_library as ctl
from splink.duckdb.blocking_rule_library import block_on
from splink.datasets import splink_datasets
df = splink_datasets.fake_1000
settings = {
"link_type": "dedupe_only",
"blocking_rules_to_generate_predictions": [
block_on("first_name"),
block_on("surname"),
],
"comparisons": [
ctl.name_comparison("first_name"),
ctl.name_comparison("surname"),
ctl.date_comparison("dob", cast_strings_to_date=True),
cl.exact_match("city", term_frequency_adjustments=True),
ctl.email_comparison("email", include_username_fuzzy_level=False),
],
}
linker = DuckDBLinker(df, settings)
linker.estimate_u_using_random_sampling(max_pairs=1e6)
blocking_rule_for_training = block_on(["first_name", "surname"])
linker.estimate_parameters_using_expectation_maximisation(blocking_rule_for_training, estimate_without_term_frequencies=True)
blocking_rule_for_training = block_on("dob")
linker.estimate_parameters_using_expectation_maximisation(blocking_rule_for_training, estimate_without_term_frequencies=True)
pairwise_predictions = linker.predict()
clusters = linker.cluster_pairwise_predictions_at_threshold(pairwise_predictions, 0.95)
clusters.as_pandas_dataframe(limit=5)You can see all of the interactive charts provided in Splink by checking out the Charts Gallery.
To find the best place to ask a question, report a bug or get general advice, please refer to our Contributing Guide.
To see how users are using Splink in the wild, check out the Use Cases section of the docs.
❓ Future of Government Awards 2023: Open Source Creation - Shortlisted, result to be announced shortly
🥈 Civil Service Awards 2023: Best Use of Data, Science, and Technology - Runner up
🥇 Analysis in Government Awards 2022: People's Choice Award - Winner
🥈 Analysis in Government Awards 2022: Innovative Methods - Runner up
🥇 Analysis in Government Awards 2020: Innovative Methods - Winner
🥇 MoJ Data and Analytical Services Directorate (DASD) Awards 2020: Innovation and Impact - Winner
If you use Splink in your research, we'd be grateful for a citation as follows:
@article{Linacre_Lindsay_Manassis_Slade_Hepworth_2022,
title = {Splink: Free software for probabilistic record linkage at scale.},
author = {Linacre, Robin and Lindsay, Sam and Manassis, Theodore and Slade, Zoe and Hepworth, Tom and Kennedy, Ross and Bond, Andrew},
year = 2022,
month = {Aug.},
journal = {International Journal of Population Data Science},
volume = 7,
number = 3,
doi = {10.23889/ijpds.v7i3.1794},
url = {https://ijpds.org/article/view/1794},
}We are very grateful to ADR UK (Administrative Data Research UK) for providing the initial funding for this work as part of the Data First project.
We are extremely grateful to professors Katie Harron, James Doidge and Peter Christen for their expert advice and guidance in the development of Splink. We are also very grateful to colleagues at the UK's Office for National Statistics for their expert advice and peer review of this work. Any errors remain our own.
While Splink is a standalone package, there are a number of repositories in the Splink ecosystem:
- splink_scalaudfs contains the code to generate User Defined Functions in scala which are then callable in Spark.
- splink_datasets contains datasets that can be installed automatically as a part of Splink through the In-build datasets functionality.
- splink_synthetic_data contains code to generate synthetic data.