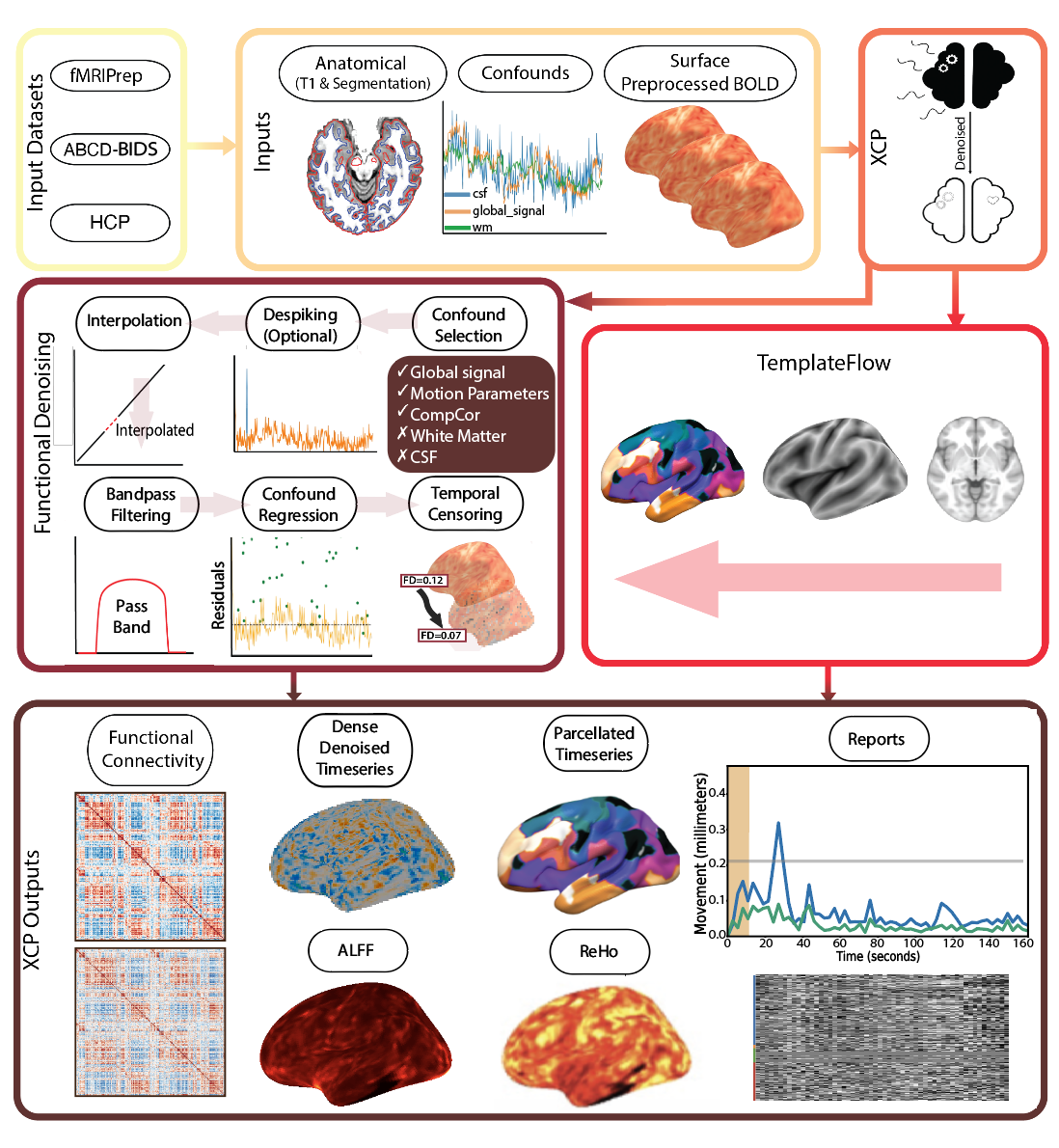
Basically, a standard cifti file run on some resting state data, just adding despiking
singularity run -e -B ${scratch}:/workdir -B ${bids_dir} $IMG ${fmriprep_dir} ${output_dir} participant --participant_label ${subject} -w /workdir -s -t rest --despike --mem_gb 24
Everything proceeds fine until reaching REHO nodes. Below is the log beginning with REHO execution:
[Node] Executing "reho_lh" <xcp_abcd.interfaces.resting_state.surfaceReho>
Downloading https://templateflow.s3.amazonaws.com/tpl-fsLR/tpl-fsLR_hemi-L_den-32k_midthickness.surf.gii
0.00B [00:00, ?B/s]
1.00B [00:00, 767B/s]211115-18:42:55,886 nipype.workflow INFO:
[Node] Finished "reho_lh", elapsed time 34.481254s.
211115-18:42:55,886 nipype.workflow WARNING:
Storing result file without outputs
211115-18:42:55,890 nipype.workflow WARNING:
[Node] Error on "xcpabcd_wf.single_subject_NDARAA306NT2_wf.cifti_postprocess_1_wf.surface_reho_wf.reho_lh" (/workdir/xcpabcd_wf/single_subject_NDARAA306NT2_wf/cifti_postprocess_1_wf/surface_reho_wf/reho_lh)
211115-18:42:57,212 nipype.workflow ERROR:
Node reho_lh failed to run on host node021.
211115-18:42:57,274 nipype.workflow ERROR:
Saving crash info to /om4/group/gablab/data/HBN/derivatives/xcp_abcd/sub-NDARAA306NT2/log/crash-20211115-184257-smeisler-reho_lh-c39954b0-7f60-4c65-83e9-16374125986d.txt
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 521, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 639, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 750, in _run_command
raise NodeExecutionError(
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node reho_lh.
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nibabel/loadsave.py", line 42, in load
stat_result = os.stat(filename)
FileNotFoundError: [Errno 2] No such file or directory: "[PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-L_den-32k_desc-vaavg_midthickness.shape.gii'), PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-L_den-32k_midthickness.surf.gii')]"
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_abcd/interfaces/resting_state.py", line 69, in _run_interface
write_gii(datat=reho_surf,template=self.inputs.surf_bold,
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_abcd/utils/write_save.py", line 133, in write_gii
template = nb.load(template)
File "/usr/local/miniconda/lib/python3.8/site-packages/nibabel/loadsave.py", line 44, in load
raise FileNotFoundError(f"No such file or no access: '{filename}'")
FileNotFoundError: No such file or no access: '[PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-L_den-32k_desc-vaavg_midthickness.shape.gii'), PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-L_den-32k_midthickness.surf.gii')]'
211115-18:42:57,365 nipype.workflow INFO:
[Node] Setting-up "xcpabcd_wf.single_subject_NDARAA306NT2_wf.cifti_postprocess_1_wf.surface_reho_wf.reho_rh" in "/workdir/xcpabcd_wf/single_subject_NDARAA306NT2_wf/cifti_postprocess_1_wf/surface_reho_wf/reho_rh".
211115-18:42:57,394 nipype.workflow INFO:
[Node] Executing "reho_rh" <xcp_abcd.interfaces.resting_state.surfaceReho>
Downloading https://templateflow.s3.amazonaws.com/tpl-fsLR/tpl-fsLR_hemi-R_den-32k_midthickness.surf.gii
0.00B [00:00, ?B/s]
1.00B [00:00, 649B/s]211115-18:43:31,806 nipype.workflow INFO:
[Node] Finished "reho_rh", elapsed time 34.404558s.
211115-18:43:31,806 nipype.workflow WARNING:
Storing result file without outputs
211115-18:43:31,832 nipype.workflow WARNING:
[Node] Error on "xcpabcd_wf.single_subject_NDARAA306NT2_wf.cifti_postprocess_1_wf.surface_reho_wf.reho_rh" (/workdir/xcpabcd_wf/single_subject_NDARAA306NT2_wf/cifti_postprocess_1_wf/surface_reho_wf/reho_rh)
211115-18:43:33,250 nipype.workflow ERROR:
Node reho_rh failed to run on host node021.
211115-18:43:33,260 nipype.workflow ERROR:
Saving crash info to /om4/group/gablab/data/HBN/derivatives/xcp_abcd/sub-NDARAA306NT2/log/crash-20211115-184333-smeisler-reho_rh-02a80c06-02a0-4fd4-9cdc-757d25e8240e.txt
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 521, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 639, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 750, in _run_command
raise NodeExecutionError(
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node reho_rh.
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nibabel/loadsave.py", line 42, in load
stat_result = os.stat(filename)
FileNotFoundError: [Errno 2] No such file or directory: "[PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-R_den-32k_desc-vaavg_midthickness.shape.gii'), PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-R_den-32k_midthickness.surf.gii')]"
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_abcd/interfaces/resting_state.py", line 69, in _run_interface
write_gii(datat=reho_surf,template=self.inputs.surf_bold,
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_abcd/utils/write_save.py", line 133, in write_gii
template = nb.load(template)
File "/usr/local/miniconda/lib/python3.8/site-packages/nibabel/loadsave.py", line 44, in load
raise FileNotFoundError(f"No such file or no access: '{filename}'")
FileNotFoundError: No such file or no access: '[PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-R_den-32k_desc-vaavg_midthickness.shape.gii'), PosixPath('/home/smeisler/.cache/templateflow/tpl-fsLR/tpl-fsLR_hemi-R_den-32k_midthickness.surf.gii')]'
211115-18:43:35,248 nipype.workflow ERROR:
could not run node: xcpabcd_wf.single_subject_NDARAA306NT2_wf.cifti_postprocess_1_wf.surface_reho_wf.reho_lh
211115-18:43:35,255 nipype.workflow ERROR:
could not run node: xcpabcd_wf.single_subject_NDARAA306NT2_wf.cifti_postprocess_1_wf.surface_reho_wf.reho_rh
Looking at my templateflow cache now, and the file appears to be there, so I'm not sure why this error is occurring. I do not get errors like this when using other BIDs apps that use templateflow.
All non-REHO outputs are present, but I'm a little confused as to why the HTMLs for the subject report no error.
I am currently running the same command but using volumetric outputs, and will update below if I get a similar error.