1King Abdullah University of Science and Technology
2NEC Laboratories China
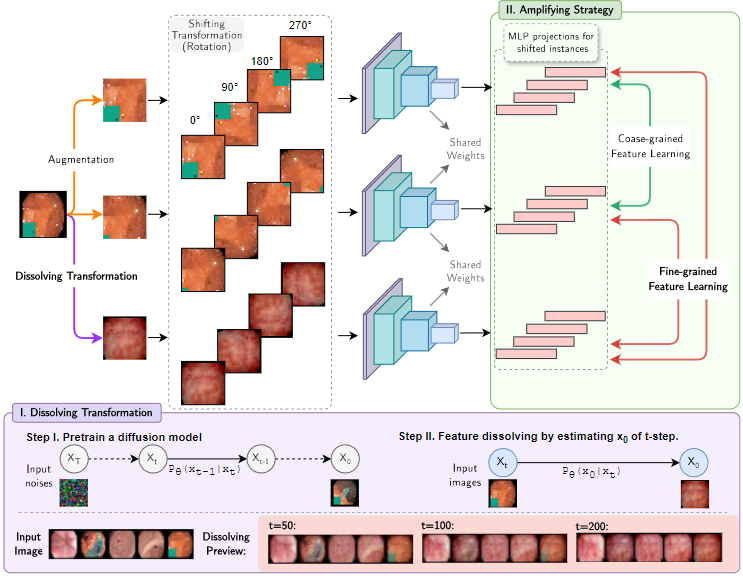
DIA is a fine-grained anomaly detection framework for medical images. We describe two novel components.
- First, the dissolving transformations. Our main observation is that generative diffusion models are feature-aware and applying them to medical images in a certain manner can remove or diminish fine-grained discriminative features such as tumors or hemorrhaging. More visual demonstration about the dissolving effects are here.
- Second, an amplifying framework. It is based on contrastive learning to learn a semantically meaningful representation of medical images in a self-supervised manner.
The amplifying framework contrasts additional pairs of images with and without dissolving transformations applied and thereby boosts the learning of fine-grained feature representations. DIA significantly improves the medical anomaly detection performance with around 18.40% AUC boost against the baseline method and achieves an overall SOTA against other benchmark methods.
$ pip install -r requirements.txt
We majorly use the following datasets to benchmark our method:
Modify the data paths in diffusion_models/train_eval_ddpm.py, then run:
python train_eval_ddpm.py --mode train --dataset <DATASET>
In our experiments, the diffusion models are usable within 5 epochs.
To train unlabeled one-class & multi-class models in the paper, run this command:
python train.py --data_root ../data --dataset <DATASET> --model resnet18_imagenet --mode simclr_DIA --shift_trans_type diffusion_rotation --diff_resolution 32 --batch_size 32 --one_class_idx 0 --save_step 1 --diffusion_model_path <DIFFUSION_MODEL_PATH>- Other transformation types are available to use, e.g.
diffusion_rotation,diffusion_cutperm,blurgaussian_rotation,blurgaussian_cutperm,blurmedian_rotation,blurmedian_cutperm. Note that cutperm may perform better if your dataset is more rotation-invariant, while the rotation may perform better if the dataset is more "permutation-invariant". - To test different resolutions, do remember to re-train the diffusion models for the corresponding resolution.
- For low resolution datasets, e.g. CIFAR10, use
--model resnet18instead of--model resnet18_imagenet.
During the evaluation, we use only rotation shift transformation for evaluation.
python eval.py --mode ood_pre --dataset <DATASET> --model resnet18_imagenet --ood_score CSI --shift_trans_type rotation --print_score --diffusion_upper_offset 0. --diffusion_offset 0. --ood_samples 10 --resize_factor 0.54 --resize_fix --one_class_idx 0 --load_path <MODEL_PATH>In general, our method achieves SOTA in terms of fine-grained anomaly detection tasks. We recommend using 32x32 diffusion_rotation for most tasks, as the following results:
In general, diffusion_rotation performs best.
| Dataset | transform | Gaussian | Median | Diffusion |
|---|---|---|---|---|
| SARS-COV-2 | Perm | 0.788 | 0.826 | 0.841 |
| Rotate | 0.847 | 0.837 | 0.851 | |
| Kvasir | Perm | 0.712 | 0.663 | 0.840 |
| Rotate | 0.739 | 0.687 | 0.860 | |
| Retinal OCT | Perm | 0.754 | 0.747 | 0.890 |
| Rotate | 0.895 | 0.876 | 0.919 | |
| APTOS 2019 | Perm | 0.942 | 0.929 | 0.926 |
| Rotate | 0.922 | 0.918 | 0.934 |
Higher feature dissolving resolution will dramatically increase the processing time, while hardly bringing up the detection performances.
| Dataset | resolution | MACs (G) | AUC |
|---|---|---|---|
| SARS-COV-2 | 32 | 2.33 | 0.851 |
| 64 | 3.84 | 0.803 | |
| 128 | 9.90 | 0.807 | |
| Kvasir | 32 | 2.33 | 0.860 |
| 64 | 3.84 | 0.721 | |
| 128 | 9.90 | 0.730 | |
| Retinal OCT | 32 | 2.33 | 0.919 |
| 64 | 3.84 | 0.922 | |
| 128 | 9.90 | 0.930 | |
| APTOS 2019 | 32 | 2.33 | 0.934 |
| 64 | 3.84 | 0.937 | |
| 128 | 9.90 | 0.905 |
@misc{2302.14696,
Author = {Jian Shi and Pengyi Zhang and Ni Zhang and Hakim Ghazzai and Peter Wonka},
Title = {Dissolving Is Amplifying: Towards Fine-Grained Anomaly Detection},
Year = {2023},
Eprint = {arXiv:2302.14696},
}
Our method is based on CSI.