Dissertation for the MSc Artificial Intelligence at the University of St Andrews (2020).
The final report can be read here: Breast Cancer Detection in Mammograms using Deep Learning Techniques, Adam Jaamour (2020)
The objective of this dissertation is to explore various deep learning techniques that can be used to implement a system which learns how to detect instances of breast cancer in mammograms. Nowadays, breast cancer claims 11,400 lives on average every year in the UK, making it one of the deadliest diseases. Mammography is the gold standard for detecting early signs of breast cancer, which can help cure the disease during its early stages. However, incorrect mammography diagnoses are common and may harm patients through unnecessary treatments and operations (or a lack of treatments). Therefore, systems that can learn to detect breast cancer on their own could help reduce the number of incorrect interpretations and missed cases.
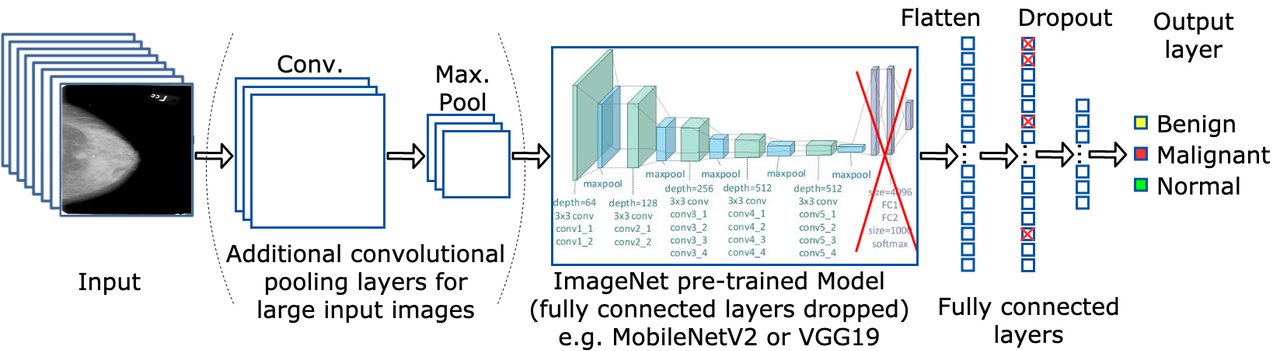
Convolution Neural Networks (CNNs) are used as part of a deep learning pipeline initially developed in a group and further extended individually. A bag-of-tricks approach is followed to analyse the effects on performance and efficiency using diverse deep learning techniques such as different architectures (VGG19, ResNet50, InceptionV3, DenseNet121, MobileNetV2), class weights, input sizes, amounts of transfer learning, and types of mammograms.
Ultimately, 67.08% accuracy is achieved on the CBIS-DDSM dataset by transfer learning pre-trained ImagetNet weights to a MobileNetV2 architecture and pre-trained weights from a binary version of the mini-MIAS dataset to the fully connected layers of the model. Furthermore, using class weights to fight the problem of imbalanced datasets and splitting CBIS-DDSM samples between masses and calcifications also increases the overall accuracy. Other techniques tested such as data augmentation and larger image sizes do not yield increased accuracies, while the mini-MIAS dataset proves to be too small for any meaningful results using deep learning techniques. These results are compared with other papers using the CBIS-DDSM and mini-MIAS datasets, and with the baseline set during the implementation of a deep learning pipeline developed as a group.
Clone the repository:
cd ~/Projects
git clone https://github.com/Adamouization/Breast-Cancer-Detection-Code
Create a repository that will be used to install Tensorflow 2 with CUDA 10 for Python and activate the virtual environment for GPU usage:
cd libraries/tf2
tar xvzf tensorflow2-cuda-10-1-e5bd53b3b5e6.tar.gz
sh build.sh
Activate the virtual environment:
source /cs/scratch/<username>/tf2/venv/bin/activate
Create outputand save_models directories to store the results:
mkdir output
mkdir saved_models
cd into the src directory and run the code:
main.py [-h] -d DATASET [-mt MAMMOGRAMTYPE] -m MODEL [-r RUNMODE] [-lr LEARNING_RATE] [-b BATCHSIZE] [-e1 MAX_EPOCH_FROZEN] [-e2 MAX_EPOCH_UNFROZEN] [-roi] [-v] [-n NAME]
where:
-his a flag for help on how to run the code.DATASETis the dataset to use. Must be eithermini-MIAS,mini-MIAS-binaryorCBIS-DDMS. Defaults toCBIS-DDMS.MAMMOGRAMTYPEis the type of mammograms to use. Can be eithercalc,massorall. Defaults toall.MODELis the model to use. Must be eitherVGG-common,VGG,ResNet,Inception,DenseNet,MobileNetorCNN.RUNMODEis the mode to run in (trainortest). Default value istrain.LEARNING_RATEis the optimiser's initial learning rate when training the model during the first training phase (frozen layers). Defaults to0.001. Must be a positive float.BATCHSIZEis the batch size to use when training the model. Defaults to2. Must be a positive integer.MAX_EPOCH_FROZENis the maximum number of epochs in the first training phrase (with frozen layers). Defaults to100.MAX_EPOCH_UNFROZENis the maximum number of epochs in the second training phrase (with unfrozen layers). Defaults to50.-roiis a flag to use versions of the images cropped around the ROI. Only usable with mini-MIAS dataset. Defaults toFalse.-vis a flag controlling verbose mode, which prints additional statements for debugging purposes.NAMEis name of the experiment being tested (used for saving plots and model weights). Defaults to an empty string.
-
This example will use the mini-MIAS dataset. After cloning the project, travel to the
data/mini-MIASdirectory (there should be 3 files in it). -
Create
images_originalandimages_processeddirectories in this directory:
cd data/mini-MIAS/
mkdir images_original
mkdir images_processed
- Move to the
images_originaldirectory and download the raw un-processed images:
cd images_original
wget http://peipa.essex.ac.uk/pix/mias/all-mias.tar.gz
- Unzip the dataset then delete all non-image files:
tar xvzf all-mias.tar.gz
rm -rf *.txt
rm -rf README
- Move back up one level and move to the
images_processeddirectory. Create 3 new directories there (benign_cases,malignant_casesandnormal_cases):
cd ../images_processed
mkdir benign_cases
mkdir malignant_cases
mkdir normal_cases
- Now run the python script for processing the dataset and render it usable with Tensorflow and Keras:
python3 ../../../src/dataset_processing_scripts/mini-MIAS-initial-pre-processing.py
These datasets are very large (exceeding 160GB) and more complex than the mini-MIAS dataset to use. They were downloaded by the University of St Andrews School of Computer Science computing officers onto \textit{BigTMP}, a 15TB filesystem that is mounted on the Centos 7 computer lab clients with NVIDIA GPUsusually used for storing large working data sets. Therefore, the download process of these datasets will not be covered in these instructions.\
The generated CSV files to use these datasets can be found in the /data/CBIS-DDSM directory, but the mammograms will have to be downloaded separately. The DDSM dataset can be downloaded here, while the CBIS-DDSM dataset can be downloaded here.
- see LICENSE file.
- Adam Jaamour
- Ashay Patel
- Shuen-Jen Chen
The common pipeline can be found at DOI 10.5281/zenodo.3975092
- Email: [email protected]
- Website: www.adam.jaamour.com
- LinkedIn: linkedin.com/in/adamjaamour