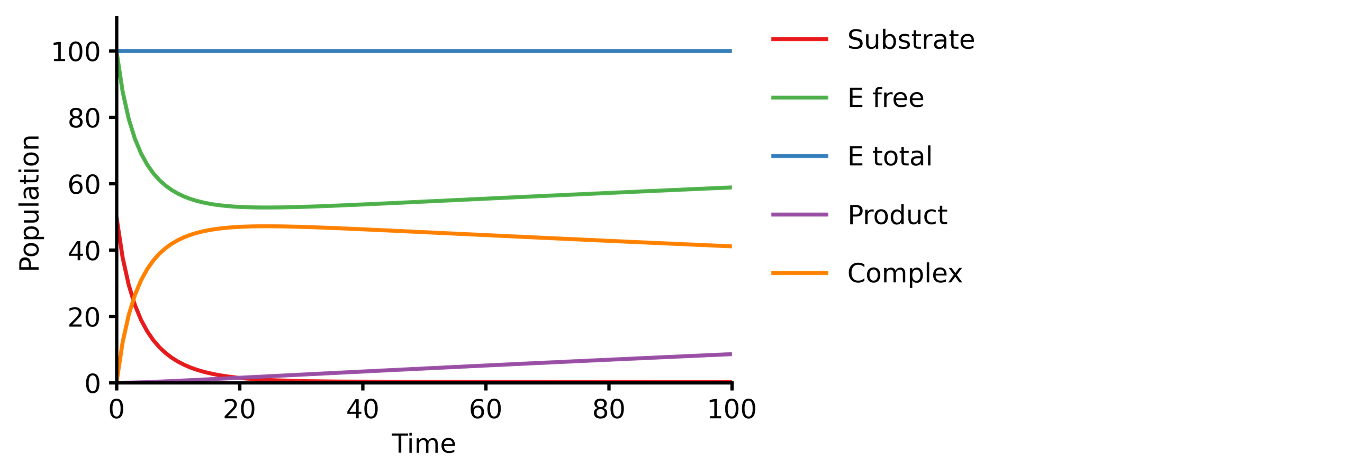This module provides a Julia interface to the BioMASS parameter estimation.
The package is a registered package, and can be installed with Pkg.add.
julia> using Pkg; Pkg.add("BioMASS")or through the pkg REPL mode by typing
] add BioMASS
- numpy - https://numpy.org
- scipy - https://scipy.org
- matplotlib - https://matplotlib.org
This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model.
E + S ⇄ ES → E + P
pasmopy.Text2Model allows you to build a BioMASS model from text. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model.
Prepare a text file describing the biochemical reactions (e.g., michaelis_menten.txt)
E + S ⇄ ES | kf=0.003, kr=0.001 | E=100, S=50
ES → E + P | kf=0.002
@obs Substrate: u[S]
@obs E_free: u[E]
@obs E_total: u[E] + u[ES]
@obs Product: u[P]
@obs Complex: u[ES]
@sim tspan: [0, 100]
Convert the text into an executable model
$ python # pasmopy requires Python 3.7+>>> from pasmopy import Text2Model
>>> description = Text2Model("michaelis_menten.txt", lang="julia")
>>> description.convert() # generate 'michaelis_menten_jl/'Simulate the model using BioMASS.jl
$ juliausing BioMASS
model = Model("./michaelis_menten_jl");
run_simulation(model)using BioMASS
model = Model("./examples/fos_model");
# Estimate unknown model parameters from experimental observations
scipy_differential_evolution(model, 1) # requires scipy package
# Save simulation results to figure/ in the model folder
run_simulation(model, viz_type="best", show_all=true)-
Imoto, H., Zhang, S. & Okada, M. A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway. Cancers 12, 2878 (2020). https://doi.org/10.3390/cancers12102878
-
Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. iScience 25, 103944 (2022). https://doi.org/10.1016/j.isci.2022.103944