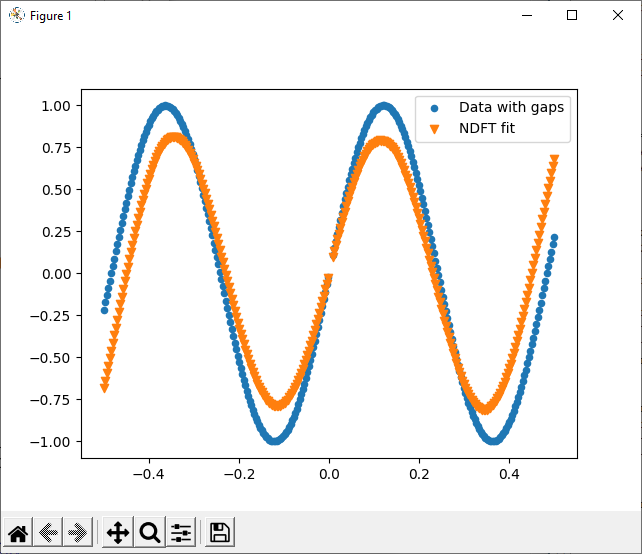
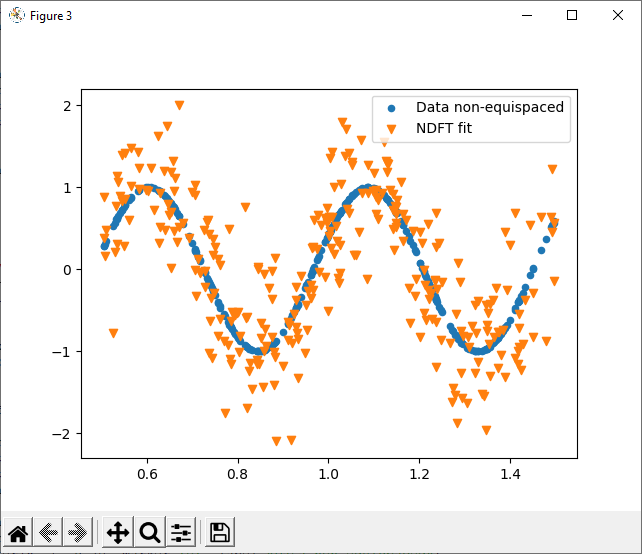
The nfft package is a lightweight implementation of the non-equispaced
fast Fourier transform (NFFT), implemented via numpy and scipy and released
under the MIT license.
For information about the NFFT algorithm, see the paper
Using NFFT 3 – a software library for various nonequispaced fast Fourier transforms.
The nfft package achieves comparable performance to the C package
described in that paper, without any customized compiled code. Rather, it
makes use of the computational building blocks available in NumPy and SciPy.
For a discussion of the algorithm and this implementation, see the
Implementation Walkthrough
notebook.
The nfft package implements one-dimensional versions of the forward and
adjoint non-equispaced fast Fourier transforms;
The forward transform:
And the adjoint transform:
In both cases, the wavenumbers k are on a regular grid from -N/2 to N/2, while the data values x_j are irregularly spaced between -1/2 and 1/2.
The direct and fast version of these algorithms are implemented in the following functions:
nfft.ndft: direct forward non-equispaced Fourier transformnfft.nfft: fast forward non-equispaced Fourier transformnfft.ndft_adjoint: direct adjoint non-equispaced Fourier transformnfft.nfft_adjoint: fast adjoint non-equispaced Fourier transform
The direct version of each transform has a computational complexity of approximately O[NM], while the NFFT has a computational complexity of approximately O[N log(N) + M log(1/ϵ)], where ϵ is the desired precision of the result. In the current implementation, memory requirements scale as approximately O[N + M log(1/ϵ)].
Another option for computing the NFFT in Python is to use the
pynfft package, which provides a
Python wrapper to the C library referenced in the above paper.
The advantage of pynfft is that, compared to nfft, it provides a more
complete set of routines, including multi-dimensional NFFTs, several related
extensions, and a range of computing strategies.
The disadvantage is that pynfft is GPL-licensed (and thus can't be used
in much of the more permissively licensed Python scientific world), and has
a much more complicated set of dependencies.
Performance-wise, nfft and pynfft are comparable, with the
implementation within nfft package being up to a factor of 2 faster
in most cases of interest (see Benchmarks.ipynb
for some simple benchmarks).
If you're curious about the implementation and how nfft attains such
performance without a custom compiled extension, see the Implementation
Walkthrough notebook.
import numpy as np
from nfft import nfft
# define evaluation points
x = -0.5 + np.random.rand(1000)
# define Fourier coefficients
N = 10000
k = - N // 2 + np.arange(N)
f_k = np.random.randn(N)
# non-equispaced fast Fourier transform
f = nfft(x, f_k)For some more examples, see the notebooks in the notebooks directory.
The nfft package can be installed directly from the Python Package Index:
$ pip install nfft
Dependencies are numpy, scipy, and pytest, and the package is tested in Python versions 2.7. 3.5, and 3.6.
Unit tests can be run using pytest:
$ pytest --pyargs nfft
This code is released under the MIT License. For more information, see the Open Source Initiative
Development of this package is supported by the UW eScience Institute, with funding from the Gordon & Betty Moore Foundation, the Alfred P. Sloan Foundation, and the Washington Research Foundation