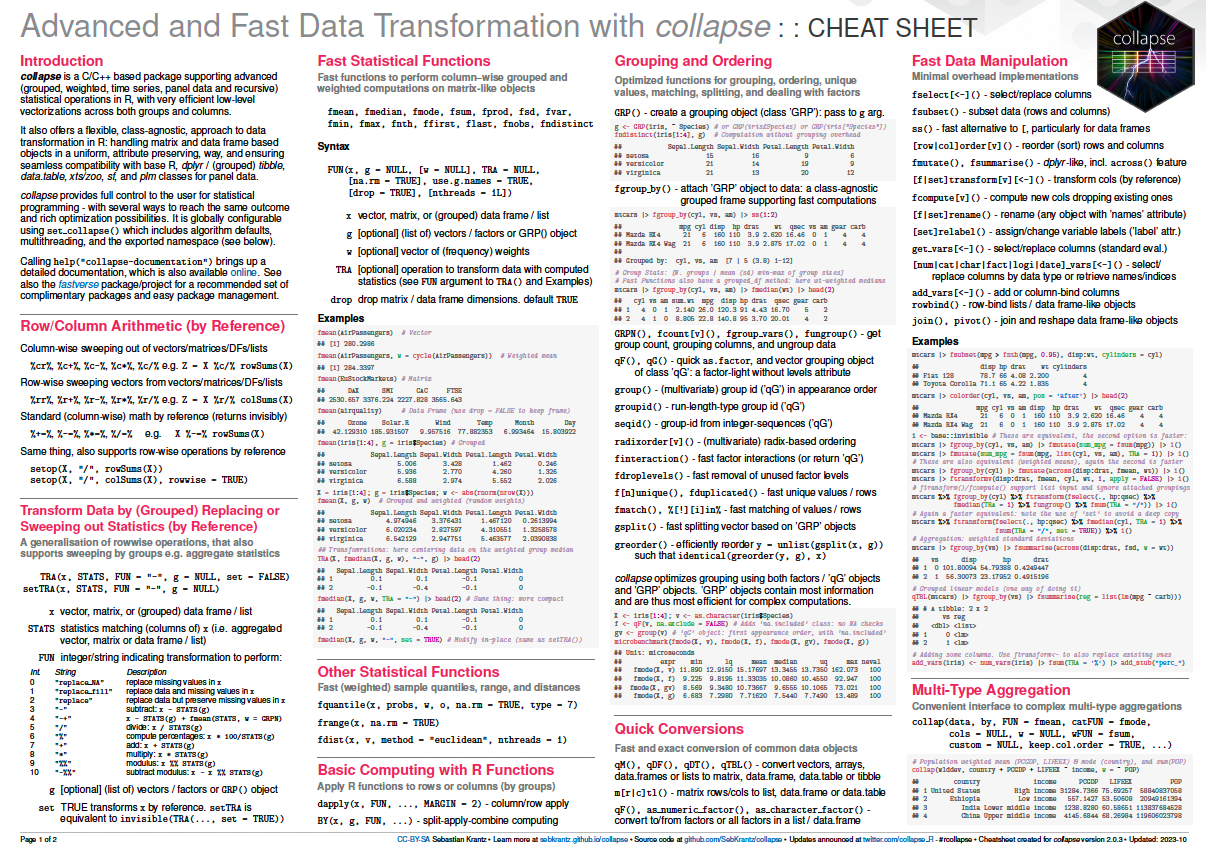
collapse is a C/C++ based package for data transformation and statistical computing in R. Its aims are:
- To facilitate complex data transformation, exploration and computing tasks in R.
- To help make R code fast, flexible, parsimonious and programmer friendly.
It further implements a class-agnostic approach to R programming, supporting base R, tibble, grouped_df (tidyverse), data.table, sf, units, pseries, pdata.frame (plm), and xts/zoo.
Key Features:
-
Advanced statistical programming: A full set of fast statistical functions supporting grouped and weighted computations on vectors, matrices and data frames. Fast and programmable grouping, ordering, matching, unique values/rows, factor generation and interactions.
-
Fast data manipulation: Fast and flexible functions for data manipulation, data object conversions, and memory efficient R programming.
-
Advanced aggregation: Fast and easy multi-data-type, multi-function, weighted and parallelized data aggregation.
-
Advanced transformations: Fast row/column arithmetic, (grouped) replacing and sweeping out of statistics (by reference), (grouped, weighted) scaling/standardizing, (higher-dimensional) between (averaging) and (quasi-)within (demeaning) transformations.
-
Advanced time-computations: Fast and flexible indexed time series and panel data classes, (sequences of) lags/leads, and (lagged/leaded, iterated, quasi-, log-) differences and (compounded) growth rates on (irregular) time series and panels. Multivariate auto-, partial- and cross-correlation functions for panel data. Panel data to (ts-)array conversions.
-
List processing: Recursive list search, splitting, extraction/subsetting, apply, and generalized row-binding / unlisting to data frame.
-
Advanced data exploration: Fast (grouped, weighted, panel-decomposed) summary statistics and descriptive tools.
collapse is written in C and C++ and only depends on Rcpp. Its algorithms are multiple times faster than base R's, scale well to <= 1 billion obs, and very efficient for complex tasks (e.g. quantiles, weighted stats, mode/counting/deduplication, joins). Optimized R code ensures minimal overheads and fast syntax evaluation.
# Install the current version on CRAN
install.packages("collapse")
# Install a stable development version (Windows/Mac binaries) from R-universe
install.packages("collapse", repos = "https://fastverse.r-universe.dev")
# Install a stable development version from GitHub (requires compilation)
remotes::install_github("SebKrantz/collapse")
# Install previous versions from the CRAN Archive (requires compilation)
install.packages("https://cran.r-project.org/src/contrib/Archive/collapse/collapse_1.9.6.tar.gz",
repos = NULL, type = "source")
# Older stable versions: 1.8.9, 1.7.6, 1.6.5, 1.5.3, 1.4.2, 1.3.2, 1.2.1collapse installs with a built-in structured documentation, implemented via a set of separate help pages. Calling help('collapse-documentation') brings up the the top-level documentation page, providing an overview of the entire package and links to all other documentation pages.
In addition there are several vignettes, among them one on Documentation and Resources.
An article on collapse has been submitted to the Journal of Statistical Software in March 2024.
Presentation at useR 2022
This provides a simple set of examples introducing some important features of collapse. It should be easy to follow for readers familiar with R.
Click here to expand
library(collapse)
data("iris") # iris dataset in base R
v <- iris$Sepal.Length # Vector
d <- num_vars(iris) # Saving numeric variables (could also be a matrix, statistical functions are S3 generic)
g <- iris$Species # Grouping variable (could also be a list of variables)
## Advanced Statistical Programming -----------------------------------------------------------------------------
# Simple (column-wise) statistics...
fmedian(v) # Vector
fsd(qM(d)) # Matrix (qM is a faster as.matrix)
fmode(d) # data.frame
fmean(qM(d), drop = FALSE) # Still a matrix
fmax(d, drop = FALSE) # Still a data.frame
# Fast grouped and/or weighted statistics
w <- abs(rnorm(fnrow(iris)))
fmedian(d, w = w) # Simple weighted statistics
fnth(d, 0.75, g) # Grouped statistics (grouped third quartile)
fmedian(d, g, w) # Groupwise-weighted statistics
fsd(v, g, w) # Similarly for vectors
fmode(qM(d), g, w, ties = "max") # Or matrices (grouped and weighted maximum mode) ...
# A fast set of data manipulation functions allows complex piped programming at high speeds
library(magrittr) # Pipe operators
iris %>% fgroup_by(Species) %>% fndistinct # Grouped distinct value counts
iris %>% fgroup_by(Species) %>% fmedian(w) # Weighted group medians
iris %>% add_vars(w) %>% # Adding weight vector to dataset
fsubset(Sepal.Length < fmean(Sepal.Length), Species, Sepal.Width:w) %>% # Fast selecting and subsetting
fgroup_by(Species) %>% # Grouping (efficiently creates a grouped tibble)
fvar(w) %>% # Frequency-weighted group-variance, default (keep.w = TRUE)
roworder(sum.w) # also saves group weights in a column called 'sum.w'
# Can also use dplyr (but dplyr manipulation verbs are a lot slower)
library(dplyr)
iris %>% add_vars(w) %>%
filter(Sepal.Length < fmean(Sepal.Length)) %>%
select(Species, Sepal.Width:w) %>%
group_by(Species) %>%
fvar(w) %>% arrange(sum.w)
## Fast Data Manipulation ---------------------------------------------------------------------------------------
head(GGDC10S)
# Pivot Wider: Only SUM (total)
SUM <- GGDC10S |> pivot(c("Country", "Year"), "SUM", "Variable", how = "wider")
head(SUM)
# Joining with data from wlddev
wlddev |>
join(SUM, on = c("iso3c" = "Country", "year" = "Year"), how = "inner")
# Recast pivoting + supplying new labels for generated columns
pivot(GGDC10S, values = 6:16, names = list("Variable", "Sectorcode"),
labels = list(to = "Sector",
new = c(Sectorcode = "GGDC10S Sector Code",
Sector = "Long Sector Description",
VA = "Value Added",
EMP = "Employment")),
how = "recast", na.rm = TRUE)
## Advanced Aggregation -----------------------------------------------------------------------------------------
collap(iris, Sepal.Length + Sepal.Width ~ Species, fmean) # Simple aggregation using the mean..
collap(iris, ~ Species, list(fmean, fmedian, fmode)) # Multiple functions applied to each column
add_vars(iris) <- w # Adding weights, return in long format..
collap(iris, ~ Species, list(fmean, fmedian, fmode), w = ~ w, return = "long")
# Generate some additional logical data
settransform(iris, AWMSL = Sepal.Length > fmedian(Sepal.Length, w = w),
AWMSW = Sepal.Width > fmedian(Sepal.Width, w = w))
# Multi-type data aggregation: catFUN applies to all categorical columns (here AMWSW)
collap(iris, ~ Species + AWMSL, list(fmean, fmedian, fmode),
catFUN = fmode, w = ~ w, return = "long")
# Custom aggregation gives the greatest possible flexibility: directly mapping functions to columns
collap(iris, ~ Species + AWMSL,
custom = list(fmean = 2:3, fsd = 3:4, fmode = "AWMSL"), w = ~ w,
wFUN = list(fsum, fmin, fmax), # Here also aggregating the weight vector with 3 different functions
keep.col.order = FALSE) # Column order not maintained -> grouping and weight variables first
# Can also use grouped tibble: weighted median for numeric, weighted mode for categorical columns
iris %>% fgroup_by(Species, AWMSL) %>% collapg(fmedian, fmode, w = w)
## Advanced Transformations -------------------------------------------------------------------------------------
# All Fast Statistical Functions have a TRA argument, supporting 10 different replacing and sweeping operations
fmode(d, TRA = "replace") # Replacing values with the mode
fsd(v, TRA = "/") # dividing by the overall standard deviation (scaling)
fsum(d, TRA = "%") # Computing percentages
fsd(d, g, TRA = "/") # Grouped scaling
fmin(d, g, TRA = "-") # Setting the minimum value in each species to 0
ffirst(d, g, TRA = "%%") # Taking modulus of first value in each species
fmedian(d, g, w, "-") # Groupwise centering by the weighted median
fnth(d, 0.95, g, w, "%") # Expressing data in percentages of the weighted species-wise 95th percentile
fmode(d, g, w, "replace", # Replacing data by the species-wise weighted minimum-mode
ties = "min")
# TRA() can also be called directly to replace or sweep with a matching set of computed statistics
TRA(v, sd(v), "/") # Same as fsd(v, TRA = "/")
TRA(d, fmedian(d, g, w), "-", g) # Same as fmedian(d, g, w, "-")
TRA(d, BY(d, g, quantile, 0.95), "%", g) # Same as fnth(d, 0.95, g, TRA = "%") (apart from quantile algorithm)
# For common uses, there are some faster and more advanced functions
fbetween(d, g) # Grouped averaging [same as fmean(d, g, TRA = "replace") but faster]
fwithin(d, g) # Grouped centering [same as fmean(d, g, TRA = "-") but faster]
fwithin(d, g, w) # Grouped and weighted centering [same as fmean(d, g, w, "-")]
fwithin(d, g, w, theta = 0.76) # Quasi-centering i.e. d - theta*fbetween(d, g, w)
fwithin(d, g, w, mean = "overall.mean") # Preserving the overall weighted mean of the data
fscale(d) # Scaling and centering (default mean = 0, sd = 1)
fscale(d, mean = 5, sd = 3) # Custom scaling and centering
fscale(d, mean = FALSE, sd = 3) # Mean preserving scaling
fscale(d, g, w) # Grouped and weighted scaling and centering
fscale(d, g, w, mean = "overall.mean", # Setting group means to overall weighted mean,
sd = "within.sd") # and group sd's to fsd(fwithin(d, g, w), w = w)
get_vars(iris, 1:2) # Use get_vars for fast selecting data.frame columns, gv is shortcut
fhdbetween(gv(iris, 1:2), gv(iris, 3:5)) # Linear prediction with factors and continuous covariates
fhdwithin(gv(iris, 1:2), gv(iris, 3:5)) # Linear partialling out factors and continuous covariates
# This again opens up new possibilities for data manipulation...
iris %>%
ftransform(ASWMSL = Sepal.Length > fmedian(Sepal.Length, Species, w, "replace")) %>%
fgroup_by(ASWMSL) %>% collapg(w = w, keep.col.order = FALSE)
iris %>% fgroup_by(Species) %>% num_vars %>% fwithin(w) # Weighted demeaning
## Time Series and Panel Series ---------------------------------------------------------------------------------
flag(AirPassengers, -1:3) # A sequence of lags and leads
EuStockMarkets %>% # A sequence of first and second seasonal differences
fdiff(0:1 * frequency(.), 1:2)
fdiff(EuStockMarkets, rho = 0.95) # Quasi-difference [x - rho*flag(x)]
fdiff(EuStockMarkets, log = TRUE) # Log-difference [log(x/flag(x))]
EuStockMarkets %>% fgrowth(c(1, frequency(.))) # Ordinary and seasonal growth rate
EuStockMarkets %>% fgrowth(logdiff = TRUE) # Log-difference growth rate [log(x/flag(x))*100]
# Creating panel data
pdata <- EuStockMarkets %>% list(`A` = ., `B` = .) %>%
unlist2d(idcols = "Id", row.names = "Time")
L(pdata, -1:3, ~Id, ~Time) # Sequence of fully identified panel-lags (L is operator for flag)
pdata %>% fgroup_by(Id) %>% flag(-1:3, Time) # Same thing..
# collapse also supports indexed series and data frames (and plm panel data classes)
pdata <- findex_by(pdata, Id, Time)
L(pdata, -1:3) # Same as above, ...
psacf(pdata) # Multivariate panel-ACF
psmat(pdata) %>% plot # 3D-array of time series from panel data + plotting
HDW(pdata) # This projects out id and time fixed effects.. (HDW is operator for fhdwithin)
W(pdata, effect = "Id") # Only Id effects.. (W is operator for fwithin)
## List Processing ----------------------------------------------------------------------------------------------
# Some nested list of heterogenous data objects..
l <- list(a = qM(mtcars[1:8]), # Matrix
b = list(c = mtcars[4:11], # data.frame
d = list(e = mtcars[2:10],
f = fsd(mtcars)))) # Vector
ldepth(l) # List has 4 levels of nesting (considering that mtcars is a data.frame)
is_unlistable(l) # Can be unlisted
has_elem(l, "f") # Contains an element by the name of "f"
has_elem(l, is.matrix) # Contains a matrix
get_elem(l, "f") # Recursive extraction of elements..
get_elem(l, c("c","f"))
get_elem(l, c("c","f"), keep.tree = TRUE)
unlist2d(l, row.names = TRUE) # Intelligent recursive row-binding to data.frame
rapply2d(l, fmean) %>% unlist2d # Taking the mean of all elements and repeating
# Application: extracting and tidying results from (potentially nested) lists of model objects
list(mod1 = lm(mpg ~ carb, mtcars),
mod2 = lm(mpg ~ carb + hp, mtcars)) %>%
lapply(summary) %>%
get_elem("coef", regex = TRUE) %>% # Regular expression search and extraction
unlist2d(idcols = "Model", row.names = "Predictor")
## Summary Statistics -------------------------------------------------------------------------------------------
irisNA <- na_insert(iris, prop = 0.15) # Randmonly set 15% missing
fnobs(irisNA) # Observation count
pwnobs(irisNA) # Pairwise observation count
fnobs(irisNA, g) # Grouped observation count
fndistinct(irisNA) # Same with distinct values... (default na.rm = TRUE skips NA's)
fndistinct(irisNA, g)
descr(iris) # Detailed statistical description of data
varying(iris, ~ Species) # Show which variables vary within Species
varying(pdata) # Which are time-varying ?
qsu(iris, w = ~ w) # Fast (one-pass) summary (with weights)
qsu(iris, ~ Species, w = ~ w, higher = TRUE) # Grouped summary + higher moments
qsu(pdata, higher = TRUE) # Panel-data summary (between and within entities)
pwcor(num_vars(irisNA), N = TRUE, P = TRUE) # Pairwise correlations with p-value and observations
pwcor(W(pdata, keep.ids = FALSE), P = TRUE) # Within-correlations
Evaluated and more extensive sets of examples are provided on the package page (also accessible from R by calling example('collapse-package')), and further in the vignettes and documentation.
If collapse was instrumental for your research project, please consider citing it using citation("collapse").