License

Tests Windows
SnakeCharm plugin for PyCharm / IntelliJ Platform IDEs adds IDE support for Snakemake workflows language, that is widely used in Bioinformatics. The plugin is developed by JetBrains Research Team, for more details see project home page.
Please report features suggestions or found bugs to project issue tracker.
Features highlights:
NB: To activate all features please enable
Snakemakesupport inSettings | Languages & Frameworks | Snakemake. Snakemake framework should be configured to use python interpreter withsnakemakemodule installed.
Features available in Snakefile and *.smk, *.rule, *.rules files:
-
Python code syntax highlighting
-
Snakemake specific syntax highlighting
- Highlight rule section names
- Syntax highlighting for Snakemake string format language, e.g. in strings like
"results/sample_{genome}.bam". - Special highlighting for rule wildcards and their usages
-
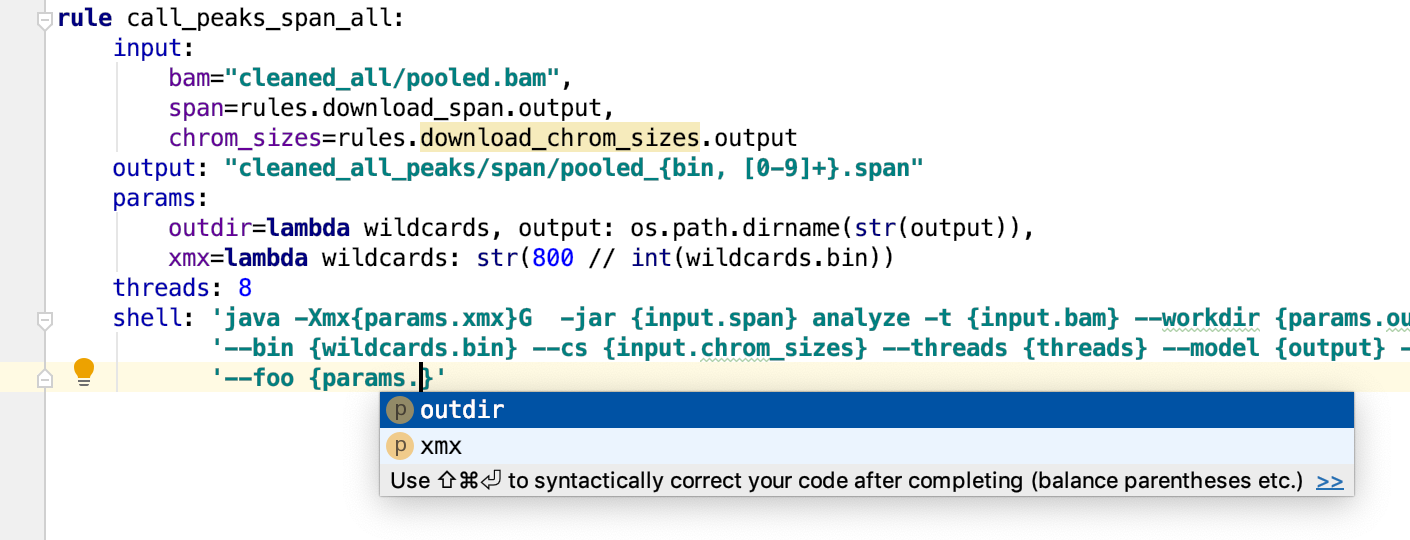
Code completion and goto to declaration for:
- Snakemake specific variables (e.g.
config,rules, etc); - Snakemake api methods like
expand,temp, etc. - Rules names after
rules., checkpoints aftercheckpoints. - Rules sections names after
rules.my_rule_name. - Rule sections keywords arguments, e.g. after
rules.my_rule_name.input. - Wildcards and sections names in
shell,input,message,paramsand other sections - Paths in sections:
configfile,workdir,conda,include. - Completion for wrappers names in
wrapper:sections - Wrappers with detailed
meta.yamlprovides completion for rule section keyword arguments inoutput,input,paramssections - Python methods and variables
- Snakemake specific variables (e.g.
-
Structure view for Snakemake files
-
Rules blocks folding
-
Open rule declaration by name using
Navigate|Symbol... -
Quick documentation for wrappers, that includes links to wrapper source code and paga in wrappers repository
-
Multiple code inspections for possible errors in snakemake files
Useful links:
Via JetBrains Plugins Market Place (recommended):
NB: If you've already configured custom file type for
Snakefileor*.smkfiles please delete it (or change file masks to smth else). Otherwise, your custom file type will replace SnakeCharm support in snakemake files.
In PyCharm (or other IntelliJ Platfrom based IDE with Python Community Edition plugin installed) open Preferences|Plugins|Marketplace|SnakeCharm and press Install button.
From ZIP plugin bundle (alternative way):
- Open IDEA/PyCharm Preferences
- Choose
Pluginssection - Press gear icon and choose
Install Plugin from Disk... - Use
*.zipbundle download from Plugin Manager or built from sources
To enable all code insight features (e.g. code completion for directory, touch etc methods) please configure Python SDK with installed snakemake package.
https://github.com/JetBrains-Research/snakecharm/wiki#setup-snakemake-support
Plugin is available in IntelliJ Plugins Repository, see SnakeCharm Plugin.
Early builds of coming releases are available in EAP channel. To receive this updates add EAP repository:
- Open
PyCharm/IntelliJ IDEAIDE settings - In Plugins section press ⚙️ (
gearicon), selectManage Plugin Repositories... - Add repo
https://plugins.jetbrains.com/plugins/eap/listusing+button, press OK (Complete guide at Custom Release Channels) - Switch to
Marketplacetab, search forsnakecharm - EAP builds will be visible as plugin updates if compatible with your IDE build
Contact me at [email protected] or post issues in Issue Tracker
Pull requests are welcome. It is my side project, so I appreciate your help with implementation of desired features. See DEVELPER.md