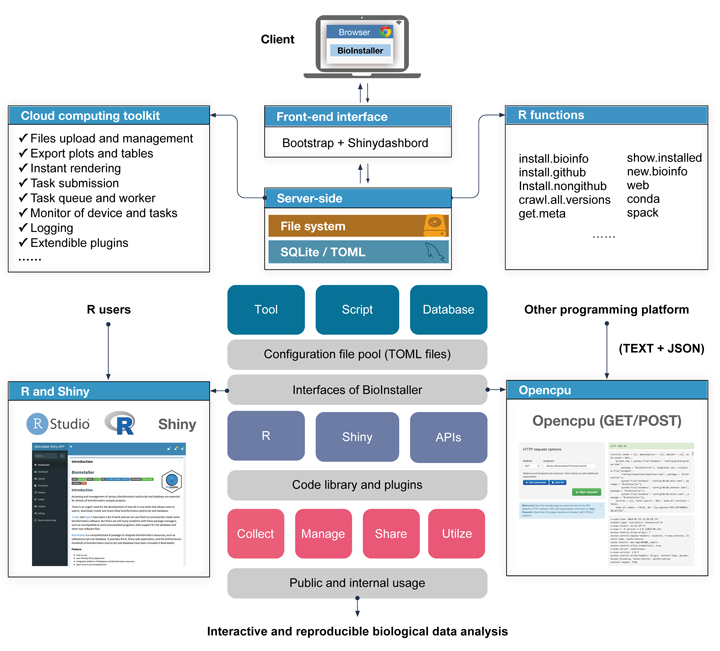
The increase in bioinformatics resources such as tools/scripts and databases poses a great challenge for users seeking to construct interactive and reproducible biological data analysis applications.
R language, as the most popular programming language for statistics, biological data analysis, and big data, has enabled diverse and free R packages (>14000) for different types of applications. However, due to the lack of high-performance and open-source cloud platforms based on R (e.g., Galaxy for Python users), it is still difficult for R users, especially those without web development skills, to construct interactive and reproducible biological data analysis applications supporting the upload and management of files, long-time computation, task submission, tracking of output files, exception handling, logging, export of plots and tables, and extendible plugin systems.
The collection, management, and share of various bioinformatics tools/scripts and databases are also essential for almost all bioinformatics analysis projects.
Here, we established a new platform to construct interactive and reproducible biological data analysis applications based on R language. This platform contains diverse user interfaces, including the R functions and R Shiny application, REST APIs, and support for collecting, managing, sharing, and utilizing massive bioinformatics tools/scripts and databases.
Feature:
- Easy-to-use
- User-friendly Shiny application
- Integrative platform of Databases and bioinformatics resources
- Open source and completely free
- One-click to download and install bioinformatics resources (via R, Shiny or Opencpu REST APIs)
- More attention for those software and database resource that have not been by other tools
- Logging
- System monitor
- Task submitting system
- Parallel tasks
Field
- Quality Control
- Alignment And Assembly
- Alternative Splicing
- ChIP-seq analysis
- Gene Expression Data Analysis
- Variant Detection
- Variant Annotation
- Virus Related
- Statistical and Visualization
- Noncoding RNA Related Database
- Cancer Genomics Database
- Regulator Related Database
- eQTL Related Database
- Clinical Annotation
- Drugs Database
- Proteomic Database
- Software Dependence Database
- ......
Note: We are developing bget and bioshiny projects independently for simplify the functions of download and shiny.
- bget is an golang-based command-line tool that do not need to install any R packages.
- bioshiny is the core shiny application of previous BioInstaller package.
#You can install this package directly from CRAN by running (from within R):
install.packages('BioInstaller')# install.packages("devtools")
devtools::install_github("JhuangLab/BioInstaller")
Note, the Shiny application of BioInstaller was migrated to bioshiny project. All shiny files in this package have been removed for reducing package size.
In the new project, we are developing more free plugins of bioshiny for various bioinformatics data analysis.
echo 'export BIO_SOFTWARES_DB_ACTIVE="~/.bioshiny/info.yaml" >> ~/.bashrc'
echo 'export BIOSHINY_CONFIG="~/.bioshiny/shiny.config.yaml" >> ~/.bashrc'
. ~/.bashrc
# Start the standalone Shiny application
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_deps_r
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_start
chmod a+x bioshiny_deps_r
chmod a+x bioshiny_start
./bioshiny_deps_r
# Start Shiny application workers
Rscript -e "bioshiny::set_shiny_workers(1)"
./bioshiny_start
# or use yarn
yarn global add bioshiny
bioshiny_deps_r
Rscript -e "bioshiny::set_shiny_workers(1)"
bioshiny_startspack and miniconda are required for extra functions.
- GitHub resource
- GitHub resource meta information
- Non GitHub resource
- Non Github resource meta infrmation
- Database
- Web Service
- Docker
Quality Control:
- FastQC, PRINSEQ, SolexaQA, FASTX-Toolkit ...
Alignment and Assembly:
- BWA, STAR, TMAP, Bowtie, Bowtie2, tophat2, hisat2, GMAP-GSNAP, ABySS, SSAHA2, Velvet, Edean, Trinity, oases, RUM, MapSplice2, NovoAlign ...
Variant Detection:
- GATK, Mutect, VarScan2, FreeBayes, LoFreq, TVC, SomaticSniper, Pindel, Delly, BreakDancer, FusionCatcher, Genome STRiP, CNVnator, CNVkit, SpeedSeq ...
Variant Annotation:
- ANNOVAR, SnpEff, VEP, oncotator ...
Utils:
- htslib, samtools, bcftools, bedtools, bamtools, vcftools, sratools, picard, HTSeq, seqtk, UCSC Utils(blat, liftOver), bamUtil, jvarkit, bcl2fastq2, fastq_tools ...
Genome:
- hisat2_reffa, ucsc_reffa, ensemble_reffa ...
Others:
- sparsehash, SQLite, pigz, lzo, lzop, bzip2, zlib, armadillo, pxz, ROOT, curl, xz, pcre, R, gatk_bundle, ImageJ, igraph ...
Databases:
- ANNOVAR, blast, CSCD, GATK_Bundle, biosystems, civic, denovo_db, dgidb, diseaseenhancer, drugbank, ecodrug, expression_atlas, funcoup, gtex, hpo, inbiomap, interpro, medreaders, mndr, msdd, omim, pancanqtl, proteinatlas, remap2, rsnp3, seecancer, srnanalyzer, superdrug2, tumorfusions, varcards ...
You can use the BioInstaller in Docker since v0.3.0. Shiny application was supported since v0.3.5.
docker pull bioinstaller/bioinstaller
docker run -it -p 80:80 -p 8004:8004 -v /tmp/download:/tmp/download bioinstaller/bioinstallerService list:
- localhost/ocpu/ Opencpu service
- localhost/shiny/BioInstaller Shiny service
- localhost/rstudio/ Rstudio server (opencpu/opencpu)
- Li J, Cui B, Dai Y, et al. BioInstaller: a comprehensive R package to construct interactive and reproducible biological data analysis applications based on the R platform[J]. PeerJ, 2018, 6:e5853.
Please fork the GitHub BioInstaller
repository, modify it, and
submit a pull request to us. Especialy, the files list in contributed section should be modified when you see a tool or database that not be
included in the other software warehouse.
R package:
Related Other Resources
Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License