| Master | Devel |
|---|---|
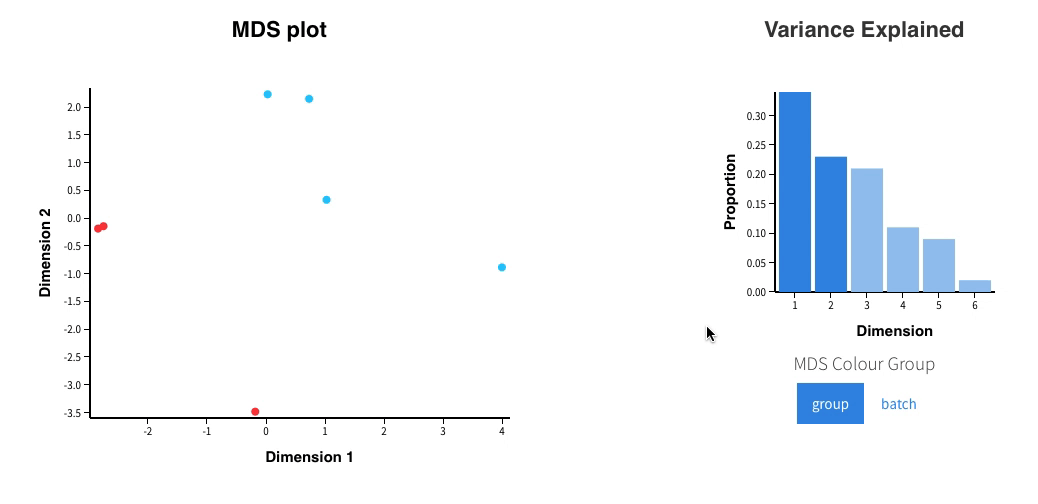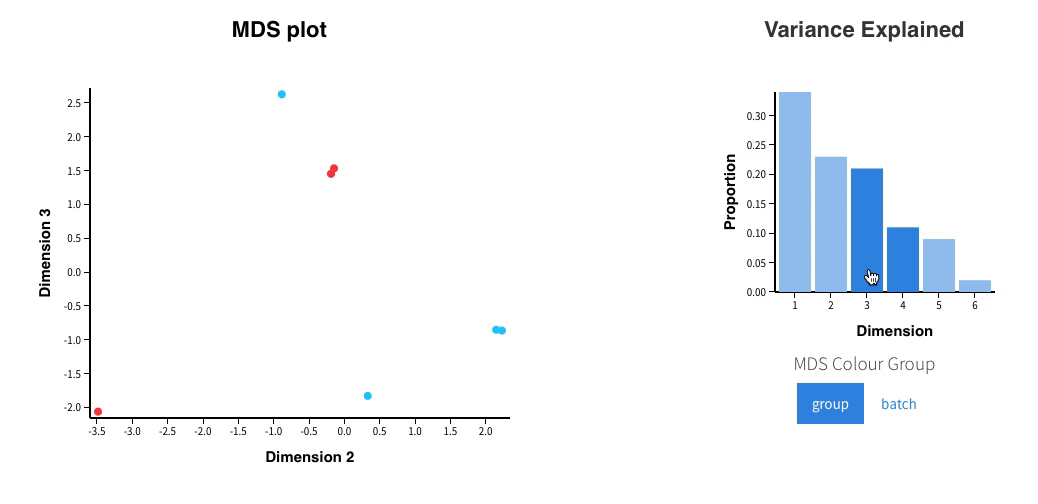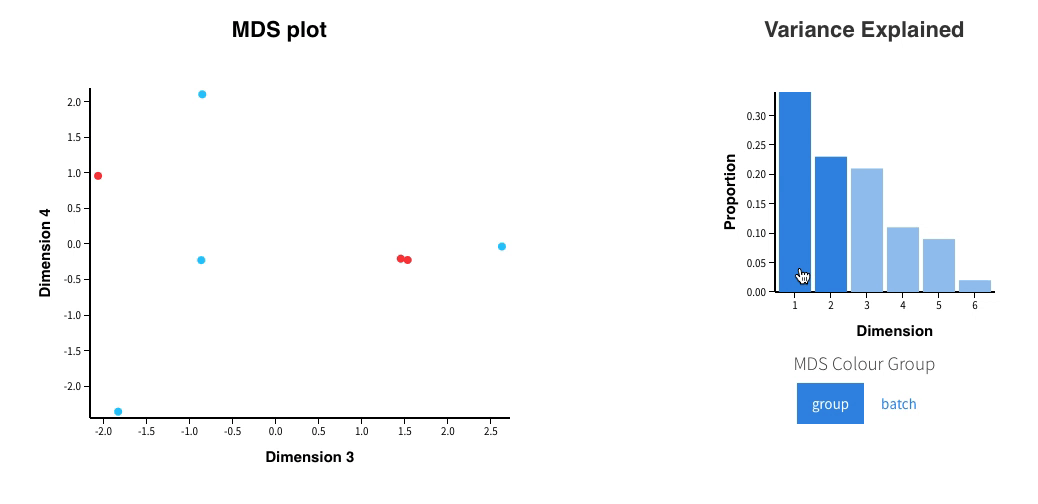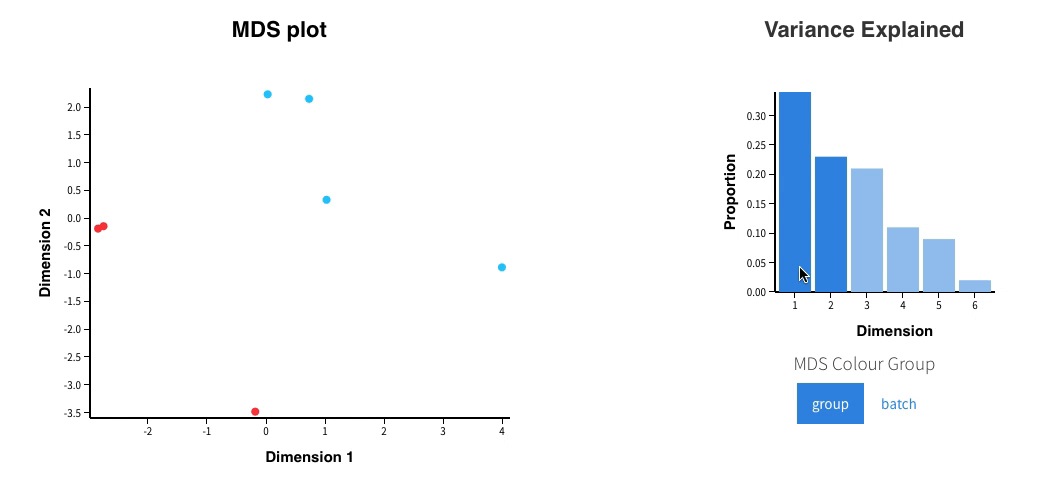
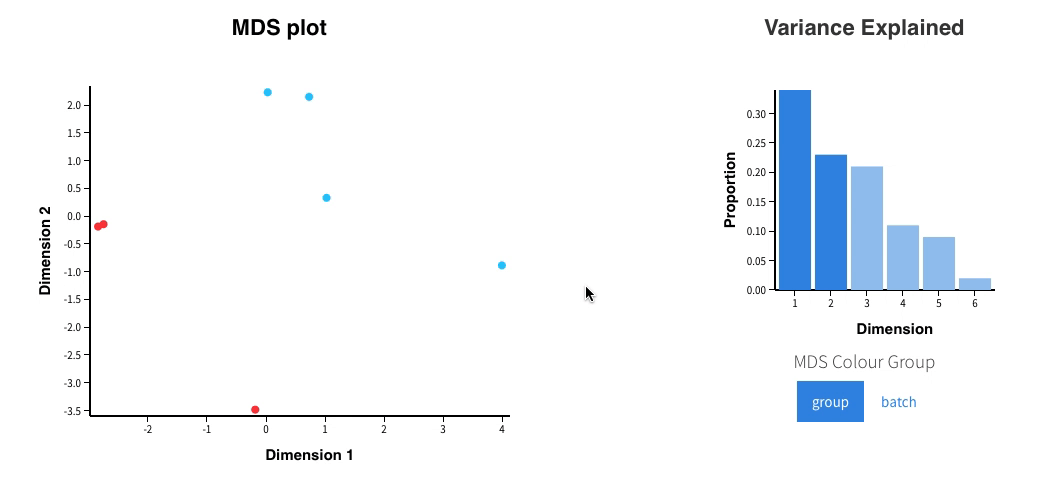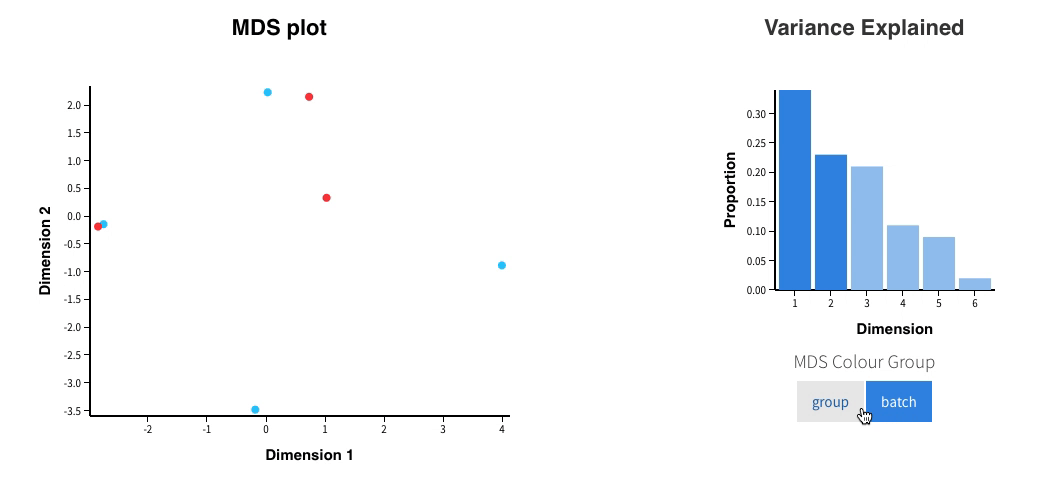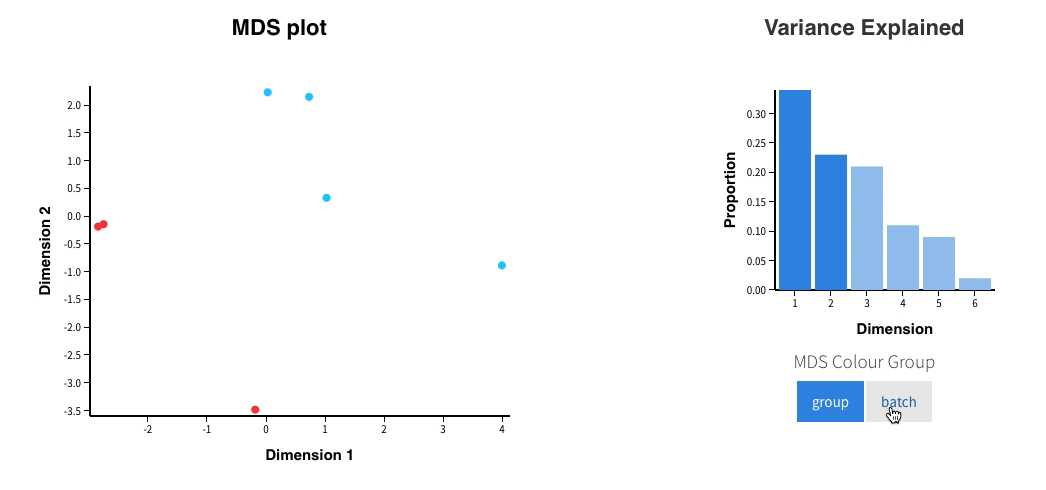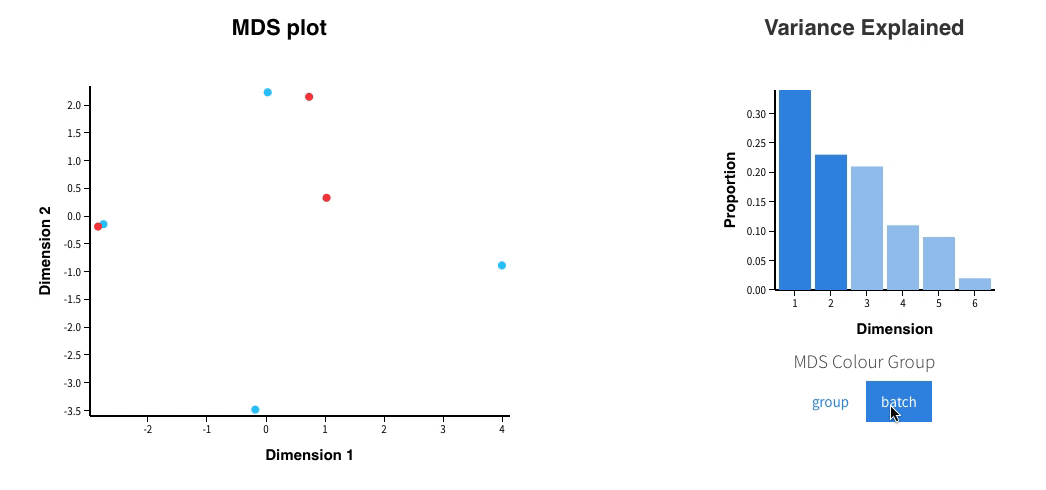
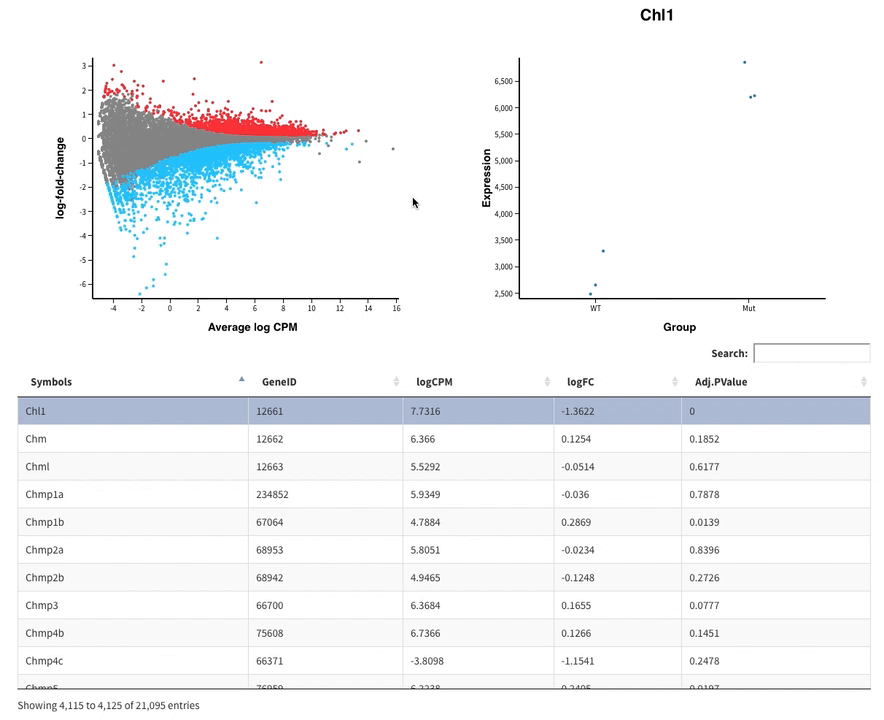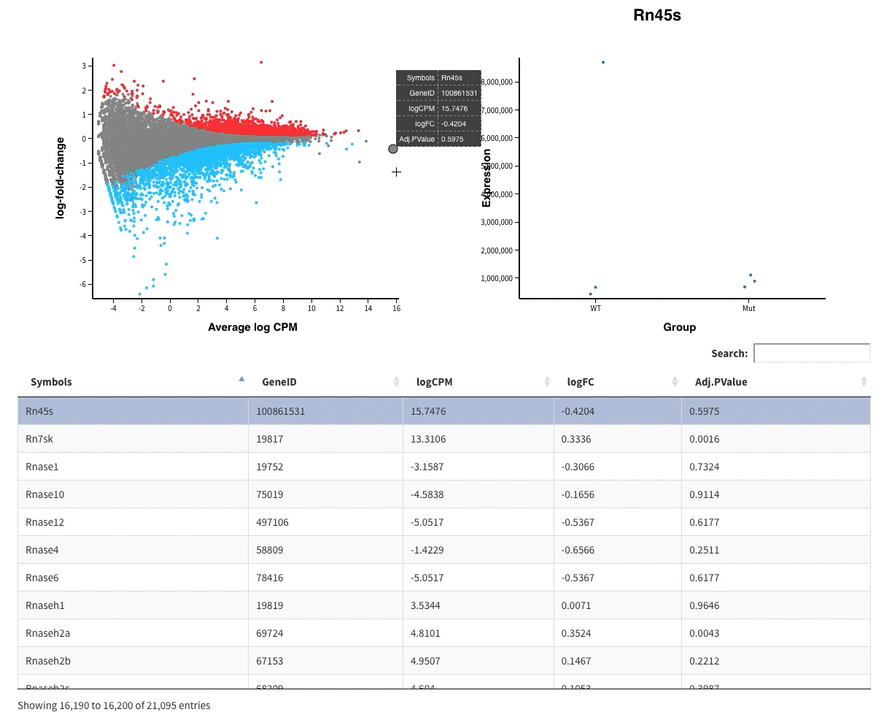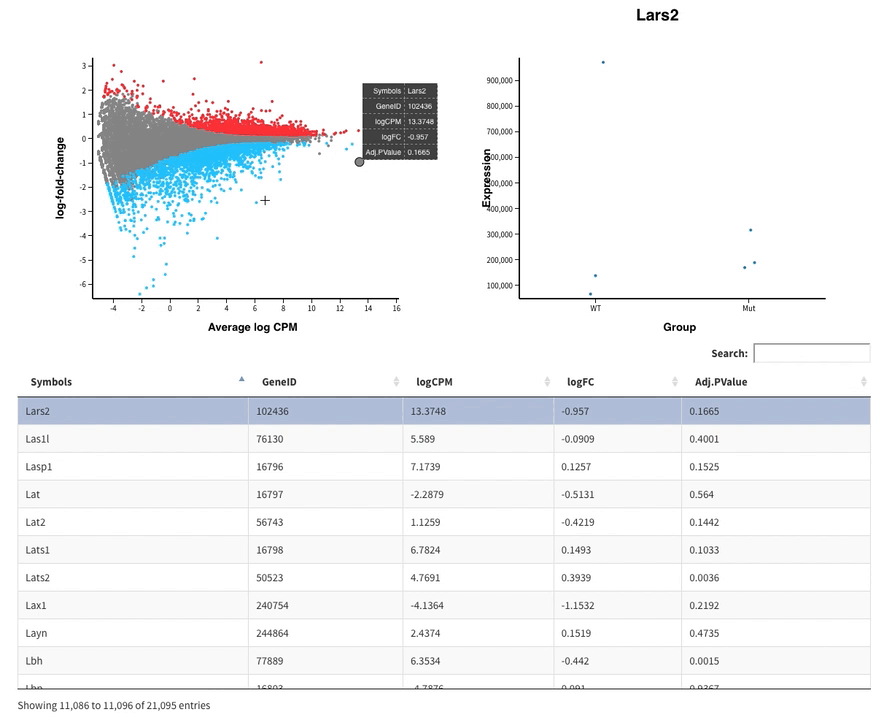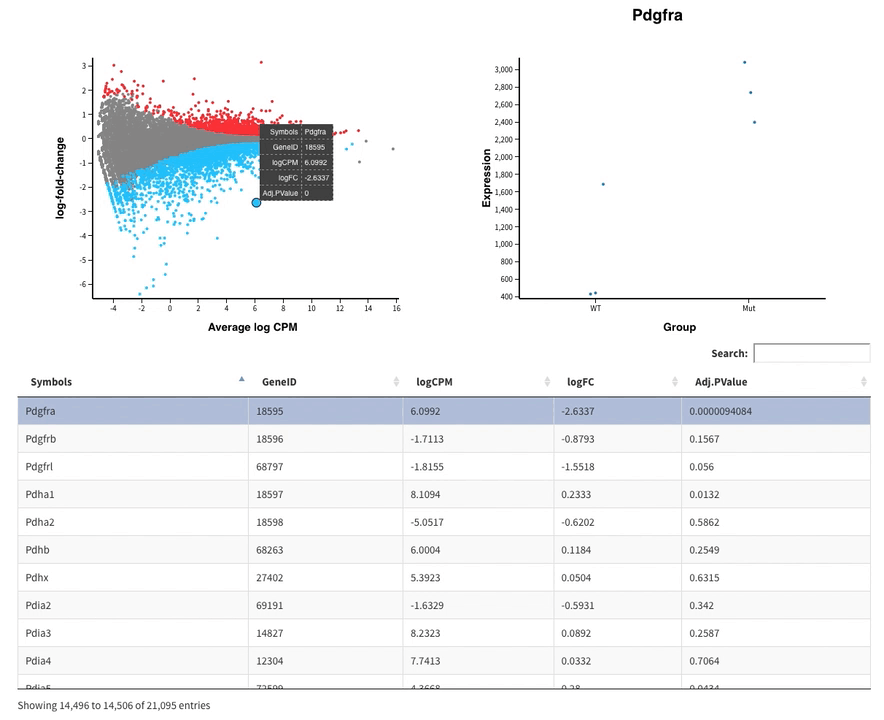
Welcome to Glimma, a R package for creating interactive plots for differential expression analysis.
User suggestions are very welcome, please start an issue for usability improvements or features you'd like to see!
This is the recommended method for installing Glimma. The release version can be installed by running the code below:
source("https://bioconductor.org/biocLite.R")
biocLite("Glimma")If a more recent version is required the developmental version:
source("https://bioconductor.org/biocLite.R")
useDevel()
biocLite("Glimma")If you find bugs please let me know by submitting an issue to this project. If possible please provide a small runnable example demonstrating the bug, operating system and R version.